Engineered resistance to Plasmodium falciparum development in transgenic Anopheles stephensi
- PMID: 21533066
- PMCID: PMC3080844
- DOI: 10.1371/journal.ppat.1002017
Engineered resistance to Plasmodium falciparum development in transgenic Anopheles stephensi
Abstract
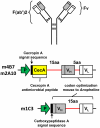
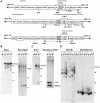
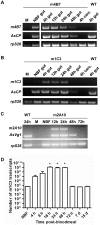
Transposon-mediated transformation was used to produce Anopheles stephensi that express single-chain antibodies (scFvs) designed to target the human malaria parasite, Plasmodium falciparum. The scFvs, m1C3, m4B7, and m2A10, are derived from mouse monoclonal antibodies that inhibit either ookinete invasion of the midgut or sporozoite invasion of salivary glands. The scFvs that target the parasite surface, m4B7 and m2A10, were fused to an Anopheles gambiae antimicrobial peptide, Cecropin A. Previously-characterized Anopheles cis-acting DNA regulatory elements were included in the transgenes to coordinate scFv production with parasite development. Gene amplification and immunoblot analyses showed promoter-specific increases in transgene expression in blood-fed females. Transgenic mosquito lines expressing each of the scFv genes had significantly lower infection levels than controls when challenged with P. falciparum.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

