In silico elucidation of the recognition dynamics of ubiquitin
- PMID: 21533067
- PMCID: PMC3080845
- DOI: 10.1371/journal.pcbi.1002035
In silico elucidation of the recognition dynamics of ubiquitin
Abstract
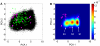
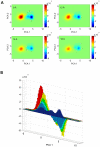
Elucidation of the mechanism of biomacromolecular recognition events has been a topic of intense interest over the past century. The inherent dynamic nature of both protein and ligand molecules along with the continuous reshaping of the energy landscape during the binding process renders it difficult to characterize this process at atomic detail. Here, we investigate the recognition dynamics of ubiquitin via microsecond all-atom molecular dynamics simulation providing both thermodynamic and kinetic information. The high-level of consistency found with respect to experimental NMR data lends support to the accuracy of the in silico representation of the conformational substates and their interconversions of free ubiquitin. Using an energy-based reweighting approach, the statistical distribution of conformational states of ubiquitin is monitored as a function of the distance between ubiquitin and its binding partner Hrs-UIM. It is found that extensive and dense sampling of conformational space afforded by the µs MD trajectory is essential for the elucidation of the binding mechanism as is Boltzmann sampling, overcoming inherent limitations of sparsely sampled empirical ensembles. The results reveal a population redistribution mechanism that takes effect when the ligand is at intermediate range of 1-2 nm from ubiquitin. This mechanism, which may be depicted as a superposition of the conformational selection and induced fit mechanisms, also applies to other binding partners of ubiquitin, such as the GGA3 GAT domain.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Fischer E. Einfluss der configuration auf die Wirkung der enzyme. Ber Dt Chem Ges. 1894;27:2985–2993.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

