Ribosomal DNA deletions modulate genome-wide gene expression: "rDNA-sensitive" genes and natural variation
- PMID: 21533076
- PMCID: PMC3080856
- DOI: 10.1371/journal.pgen.1001376
Ribosomal DNA deletions modulate genome-wide gene expression: "rDNA-sensitive" genes and natural variation
Abstract
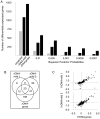
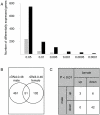
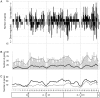
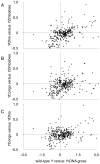
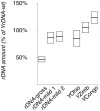
The ribosomal rDNA gene array is an epigenetically-regulated repeated gene locus. While rDNA copy number varies widely between and within species, the functional consequences of subtle copy number polymorphisms have been largely unknown. Deletions in the Drosophila Y-linked rDNA modifies heterochromatin-induced position effect variegation (PEV), but it has been unknown if the euchromatic component of the genome is affected by rDNA copy number. Polymorphisms of naturally occurring Y chromosomes affect both euchromatin and heterochromatin, although the elements responsible for these effects are unknown. Here we show that copy number of the Y-linked rDNA array is a source of genome-wide variation in gene expression. Induced deletions in the rDNA affect the expression of hundreds to thousands of euchromatic genes throughout the genome of males and females. Although the affected genes are not physically clustered, we observed functional enrichments for genes whose protein products are located in the mitochondria and are involved in electron transport. The affected genes significantly overlap with genes affected by natural polymorphisms on Y chromosomes, suggesting that polymorphic rDNA copy number is an important determinant of gene expression diversity in natural populations. Altogether, our results indicate that subtle changes to rDNA copy number between individuals may contribute to biologically relevant phenotypic variation.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
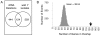
References
-
- Long EO, Dawid IB. Repeated genes in eukaryotes. Annu Rev Biochem. 1980;49:727–764. - PubMed
-
- Prokopowich CD, Gregory TR, Crease TJ. The correlation between rDNA copy number and genome size in eukaryotes. Genome. 2003;46:48–50. - PubMed
-
- Karpen GH, Schaefer JE, Laird CD. A Drosophila rRNA gene located in euchromatin is active in transcription and nucleolus formation. Genes Dev. 1988;2:1745–1763. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

