A PiggyBac-based recessive screening method to identify pluripotency regulators
- PMID: 21533166
- PMCID: PMC3078921
- DOI: 10.1371/journal.pone.0018189
A PiggyBac-based recessive screening method to identify pluripotency regulators
Abstract
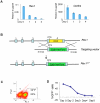
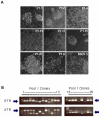
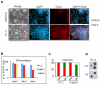
Phenotype driven genetic screens allow unbiased exploration of the genome to discover new biological regulators. Bloom syndrome gene (Blm) deficient embryonic stem (ES) cells provide an opportunity for recessive screening due to frequent loss of heterozygosity. We describe a strategy for isolating regulators of mammalian pluripotency based on conversion to homozygosity of PiggyBac gene trap insertions combined with stringent selection for differentiation resistance. From a screen of 2000 mutants we obtained a disruptive integration in the Tcf3 gene. Homozygous Tcf3 mutants showed impaired differentiation and enhanced self-renewal. This phenotype was reverted in a dosage sensitive manner by excision of one or both copies of the gene trap. These results provide new evidence confirming that Tcf3 is a potent negative regulator of pluripotency and validate a forward screening methodology to identify modulators of pluripotent stem cell biology.
Conflict of interest statement
Figures




References
-
- Luo G, Santoro IM, McDaniel LD, Nishijima I, Mills M, et al. Cancer predisposition caused by elevated mitotic recombination in Bloom mice. Nat Genet. 2000;26:424–429. - PubMed
-
- Guo G, Wang W, Bradley A. Mismatch repair genes identified using genetic screens in Blm-deficient embryonic stem cells. Nature. 2004;429:891–895. - PubMed
-
- Ding S, Wu X, Li G, Han M, Zhuang Y, et al. Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell. 2005;122:473–483. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

