A ChIP-Seq benchmark shows that sequence conservation mainly improves detection of strong transcription factor binding sites
- PMID: 21533218
- PMCID: PMC3077367
- DOI: 10.1371/journal.pone.0018430
A ChIP-Seq benchmark shows that sequence conservation mainly improves detection of strong transcription factor binding sites
Abstract
Background: Transcription factors are important controllers of gene expression and mapping transcription factor binding sites (TFBS) is key to inferring transcription factor regulatory networks. Several methods for predicting TFBS exist, but there are no standard genome-wide datasets on which to assess the performance of these prediction methods. Also, it is believed that information about sequence conservation across different genomes can generally improve accuracy of motif-based predictors, but it is not clear under what circumstances use of conservation is most beneficial.
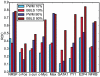
Results: Here we use published ChIP-seq data and an improved peak detection method to create comprehensive benchmark datasets for prediction methods which use known descriptors or binding motifs to detect TFBS in genomic sequences. We use this benchmark to assess the performance of five different prediction methods and find that the methods that use information about sequence conservation generally perform better than simpler motif-scanning methods. The difference is greater on high-affinity peaks and when using short and information-poor motifs. However, if the motifs are specific and information-rich, we find that simple motif-scanning methods can perform better than conservation-based methods.
Conclusions: Our benchmark provides a comprehensive test that can be used to rank the relative performance of transcription factor binding site prediction methods. Moreover, our results show that, contrary to previous reports, sequence conservation is better suited for predicting strong than weak transcription factor binding sites.
Conflict of interest statement
Figures


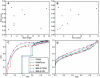
 percentile), and highest peaks (
percentile), and highest peaks ( percentile). The difference between PWM and the conservation-based BBLS PWM method is generally greater, and in favor of BBLS PWM, on the higher peaks more than the lower peaks.
percentile). The difference between PWM and the conservation-based BBLS PWM method is generally greater, and in favor of BBLS PWM, on the higher peaks more than the lower peaks.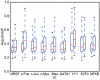
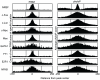
 percentile), and highest peaks (
percentile), and highest peaks ( percentile). The higher peaks generally show higher sequence conservation across genomes.
percentile). The higher peaks generally show higher sequence conservation across genomes.References
-
- Elnitski L, Jin VX, Farnham PJ, Jones SJ. Locating mammalian transcription factor binding sites: A survey of computational and experimental techniques. Genome Research. 2006;16:1455–1464. - PubMed
-
- Wasserman WW, Sandelin A. Applied bioinformatics for the identification of regulatory elements. Nat Rev Genet. 2004;5:276–287. - PubMed
-
- Tompa M, Li N, Bailey TL, Church GM, Moor BD, et al. Assessing computational tools for the discovery of transcription factor binding sites. Nat Biotech. 2005;23:137–144. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

