Improved xenobiotic metabolism and reduced susceptibility to cancer in gluten-sensitive macaques upon introduction of a gluten-free diet
- PMID: 21533263
- PMCID: PMC3075256
- DOI: 10.1371/journal.pone.0018648
Improved xenobiotic metabolism and reduced susceptibility to cancer in gluten-sensitive macaques upon introduction of a gluten-free diet
Abstract
Background: A non-human primate (NHP) model of gluten sensitivity was employed to study the gene perturbations associated with dietary gluten changes in small intestinal tissues from gluten-sensitive rhesus macaques (Macaca mulatta).
Methodology: Stages of remission and relapse were accomplished in gluten-sensitive animals by administration of gluten-free (GFD) and gluten-containing (GD) diets, as described previously. Pin-head-sized biopsies, obtained non-invasively by pediatric endoscope from duodenum while on GFD or GD, were used for preparation of total RNA and gene profiling, using the commercial Rhesus Macaque Microarray (Agilent Technologies),targeting expression of over 20,000 genes.
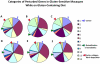
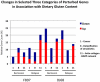
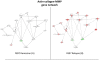
Principal findings: When compared with normal healthy control, gluten-sensitive macaques showed differential gene expressions induced by GD. While observed gene perturbations were classified into one of 12 overlapping categories--cancer, metabolism, digestive tract function, immune response, cell growth, signal transduction, autoimmunity, detoxification of xenobiotics, apoptosis, actin-collagen deposition, neuronal and unknown function--this study focused on cancer-related gene networks such as cytochrome P450 family (detoxification function) and actin-collagen-matrix metalloproteinases (MMP) genes.
Conclusions/significance: A loss of detoxification function paralleled with necessity to metabolize carcinogens was revealed in gluten-sensitive animals while on GD. An increase in cancer-promoting factors and a simultaneous decrease in cancer-preventing factors associated with altered expression of actin-collagen-MMP gene network were noted. In addition, gluten-sensitive macaques showed reduced number of differentially expressed genes including the cancer-associated ones upon withdrawal of dietary gluten. Taken together, these findings indicate potentially expanded utility of gluten-sensitive rhesus macaques in cancer research.
Conflict of interest statement
Figures





Similar articles
-
Dietary Gluten-Induced Gut Dysbiosis Is Accompanied by Selective Upregulation of microRNAs with Intestinal Tight Junction and Bacteria-Binding Motifs in Rhesus Macaque Model of Celiac Disease.Nutrients. 2016 Oct 28;8(11):684. doi: 10.3390/nu8110684. Nutrients. 2016. PMID: 27801835 Free PMC article.
-
Visualization of transepithelial passage of the immunogenic 33-residue peptide from alpha-2 gliadin in gluten-sensitive macaques.PLoS One. 2010 Apr 19;5(4):e10228. doi: 10.1371/journal.pone.0010228. PLoS One. 2010. PMID: 20419103 Free PMC article.
-
The effects of reduced gluten barley diet on humoral and cell-mediated systemic immune responses of gluten-sensitive rhesus macaques.Nutrients. 2015 Mar 6;7(3):1657-71. doi: 10.3390/nu7031657. Nutrients. 2015. PMID: 25756783 Free PMC article.
-
Exit Gluten-Free and Enter Low FODMAPs: A Novel Dietary Strategy to Reduce Gastrointestinal Symptoms in Athletes.Sports Med. 2019 Feb;49(Suppl 1):87-97. doi: 10.1007/s40279-018-01034-0. Sports Med. 2019. PMID: 30671907 Free PMC article. Review.
-
Gluten-free diet in gluten-related disorders.Dig Dis. 2013;31(1):57-62. doi: 10.1159/000347180. Epub 2013 Jun 17. Dig Dis. 2013. PMID: 23797124 Review.
Cited by
-
Gluten-sensitive enteropathy coincides with decreased capability of intestinal T cells to secrete IL-17 and IL-22 in a macaque model for celiac disease.Clin Immunol. 2013 Apr;147(1):40-49. doi: 10.1016/j.clim.2013.02.012. Epub 2013 Feb 28. Clin Immunol. 2013. PMID: 23518597 Free PMC article.
-
Role of TNF in the altered interaction of dormant Mycobacterium tuberculosis with host macrophages.PLoS One. 2014 Apr 17;9(4):e95220. doi: 10.1371/journal.pone.0095220. eCollection 2014. PLoS One. 2014. PMID: 24743303 Free PMC article.
-
Mucosal-activated invariant T cells do not exhibit significant lung recruitment and proliferation profiles in macaques in response to infection with Mycobacterium tuberculosis CDC1551.Tuberculosis (Edinb). 2019 May;116S:S11-S18. doi: 10.1016/j.tube.2019.04.006. Epub 2019 Apr 26. Tuberculosis (Edinb). 2019. PMID: 31072689 Free PMC article.
-
Microbiome signature suggestive of lactose-intolerance in rhesus macaques (Macaca mulatta) with intermittent chronic diarrhea.Anim Microbiome. 2024 Sep 23;6(1):53. doi: 10.1186/s42523-024-00338-z. Anim Microbiome. 2024. PMID: 39313845 Free PMC article.
-
Glycemic Response in Nonhuman Primates Fed Gluten-Free Rice Cakes Enriched with Soy, Pea, or Rice Protein and Its Correlation with Nutrient Composition.Nutrients. 2024 Jan 11;16(2):234. doi: 10.3390/nu16020234. Nutrients. 2024. PMID: 38257126 Free PMC article.
References
-
- Jabri B, Kasarda DD, Green PH. Innate and adaptive immunity: the yin andyang of celiac disease. Immunol Rev. 2005;206:219–31. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

