A comprehensive peptidome profiling technology for the identification of early detection biomarkers for lung adenocarcinoma
- PMID: 21533267
- PMCID: PMC3075260
- DOI: 10.1371/journal.pone.0018567
A comprehensive peptidome profiling technology for the identification of early detection biomarkers for lung adenocarcinoma
Abstract
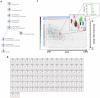
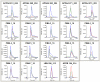
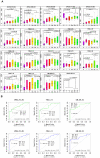
The mass spectrometry-based peptidomics approaches have proven its usefulness in several areas such as the discovery of physiologically active peptides or biomarker candidates derived from various biological fluids including blood and cerebrospinal fluid. However, to identify biomarkers that are reproducible and clinically applicable, development of a novel technology, which enables rapid, sensitive, and quantitative analysis using hundreds of clinical specimens, has been eagerly awaited. Here we report an integrative peptidomic approach for identification of lung cancer-specific serum peptide biomarkers. It is based on the one-step effective enrichment of peptidome fractions (molecular weight of 1,000-5,000) with size exclusion chromatography in combination with the precise label-free quantification analysis of nano-LC/MS/MS data set using Expressionist proteome server platform. We applied this method to 92 serum samples well-managed with our SOP (standard operating procedure) (30 healthy controls and 62 lung adenocarcinoma patients), and quantitatively assessed the detected 3,537 peptide signals. Among them, 118 peptides showed significantly altered serum levels between the control and lung cancer groups (p<0.01 and fold change >5.0). Subsequently we identified peptide sequences by MS/MS analysis and further assessed the reproducibility of Expressionist-based quantification results and their diagnostic powers by MRM-based relative-quantification analysis for 96 independently prepared serum samples and found that APOA4 273-283, FIBA 5-16, and LBN 306-313 should be clinically useful biomarkers for both early detection and tumor staging of lung cancer. Our peptidome profiling technology can provide simple, high-throughput, and reliable quantification of a large number of clinical samples, which is applicable for diverse peptidome-targeting biomarker discoveries using any types of biological specimens.
Conflict of interest statement
Figures



References
-
- Parkin DM. Global cancer statistics in the year 2000. Lancet Oncol. 2001;2:533–543. - PubMed
-
- Goldstraw P, Crowley J, Chansky K, Giroux DJ, Groome PA, et al. The IASLC Lung Cancer Staging Project: proposals for the revision of the TNM stage groupings in the forthcoming (seventh) edition of the TNM Classification of malignant tumours. J Thorac Oncol. 2007;2:706–714. - PubMed
-
- Henschke CI, Yankelevitz DF, Libby DM, Pasmantier MW, Smith JP, et al. Survival of patients with stage I lung cancer detected on CT screening. N Engl J Med. 2006;355:1763–1771. - PubMed
-
- Gail MH, Muenz L, McIntire KR, Radovich B, Braunstein G, et al. Multiple markers for lung cancer diagnosis: validation of models for localized lung cancer. J Natl Cancer Inst. 1988;80:97–101. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

