Direct interaction of FliX and FlbD is required for their regulatory activity in Caulobacter crescentus
- PMID: 21535897
- PMCID: PMC3096577
- DOI: 10.1186/1471-2180-11-89
Direct interaction of FliX and FlbD is required for their regulatory activity in Caulobacter crescentus
Abstract
Background: The temporal and spatial expression of late flagellar genes in Caulobacter crescentus is activated by the transcription factor FlbD and its partner trans-acting factor FliX. The physical interaction of these two proteins represents an alternative mechanism for regulating the activity of σ54 transcription factors. This study is to characterize the interaction of the two proteins and the consequences of the interaction on their regulatory activity.
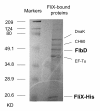
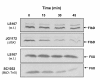
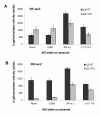
Results: FliX and FlbD form stable complexes, which can stand the interference of 2.65 M NaCl. The stability of FliX and FlbD was affected by the co-existence of each other. Five FliX mutants (R71A, L85K, Δ117-118, T130L, and L136K) were created by site-directed mutagenesis in conserved regions of the protein. All mutants were successfully expressed in both wild-type and ΔfliX Caulobacter strains. All but FliXL85K could rescue the motility and cell division defects of a ΔfliX mutant strain. The ability of FliX to regulate the transcription of class II and class III/IV flagellar promoters was fully diminished due to the L85K mutation. Co-immunoprecipitation experiment revealed that FliXL85K was unable to physically interact with FlbD.
Conclusions: FliX interacts with FlbD and thereby directly regulates the activity of FlbD in response to flagellar assembly. Mutations in highly conserved regions of FliX could severely affect the recognition between FliX and FlbD and hence interrupt the normal progression of flagellar synthesis and other developmental events in Caulobacter.
Figures





Similar articles
-
Linking structural assembly to gene expression: a novel mechanism for regulating the activity of a sigma54 transcription factor.Mol Microbiol. 2005 Nov;58(3):743-57. doi: 10.1111/j.1365-2958.2005.04857.x. Mol Microbiol. 2005. PMID: 16238624
-
Regulation of FlbD activity by flagellum assembly is accomplished through direct interaction with the trans-acting factor, FliX.Mol Microbiol. 2004 Nov;54(3):715-30. doi: 10.1111/j.1365-2958.2004.04298.x. Mol Microbiol. 2004. PMID: 15491362
-
The Caulobacter crescentus flagellar gene, fliX, encodes a novel trans-acting factor that couples flagellar assembly to transcription.Mol Microbiol. 2001 Mar;39(6):1623-37. doi: 10.1046/j.1365-2958.2001.02351.x. Mol Microbiol. 2001. PMID: 11260478
-
The role of FlbD in regulation of flagellar gene transcription in Caulobacter crescentus.Res Microbiol. 1994 Jun-Aug;145(5-6):420-30. doi: 10.1016/0923-2508(94)90090-6. Res Microbiol. 1994. PMID: 7855428 Review.
-
Cis- and trans-acting elements required for regulation of flagellar gene transcription in the bacterium Caulobacter crescentus.Cell Mol Biol Res. 1993;39(4):361-9. Cell Mol Biol Res. 1993. PMID: 8312972 Review.
Cited by
-
Genetic and Transcriptional Analyses of the Flagellar Gene Cluster in Actinoplanes missouriensis.J Bacteriol. 2016 Jul 28;198(16):2219-27. doi: 10.1128/JB.00306-16. Print 2016 Aug 15. J Bacteriol. 2016. PMID: 27274031 Free PMC article.
-
Dual Control of Flagellar Synthesis and Exopolysaccharide Production by FlbD-FliX Class II Regulatory Proteins in Bradyrhizobium diazoefficiens.J Bacteriol. 2021 Mar 8;203(7):e00403-20. doi: 10.1128/JB.00403-20. Print 2021 Mar 8. J Bacteriol. 2021. PMID: 33468586 Free PMC article.
-
Motility control through an anti-activation mechanism in Agrobacterium tumefaciens.Mol Microbiol. 2021 Nov;116(5):1281-1297. doi: 10.1111/mmi.14823. Epub 2021 Oct 19. Mol Microbiol. 2021. PMID: 34581467 Free PMC article.
References
-
- Gober JW, England J. In: Regulation of flagellum biosynthesis and motility in Caulobacter Prokaryotic Development. Brun KV, Shimkets LJ, editor. Washington, DC: American Society for Microbiology; 2000. pp. 319–339.

