Comparative genomics and the role of lateral gene transfer in the evolution of bovine adapted Streptococcus agalactiae
- PMID: 21536150
- PMCID: PMC3139733
- DOI: 10.1016/j.meegid.2011.04.019
Comparative genomics and the role of lateral gene transfer in the evolution of bovine adapted Streptococcus agalactiae
Abstract
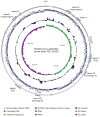
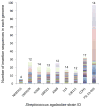
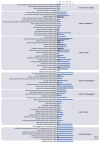
In addition to causing severe invasive infections in humans, Streptococcus agalactiae, or group B Streptococcus (GBS), is also a major cause of bovine mastitis. Here we provide the first genome sequence for S. agalactiae isolated from a cow diagnosed with clinical mastitis (strain FSL S3-026). Comparison to eight S. agalactiae genomes obtained from human disease isolates revealed 183 genes specific to the bovine strain. Subsequent polymerase chain reaction (PCR) screening for the presence/absence of a subset of these loci in additional bovine and human strains revealed strong differentiation between the two groups (Fisher exact test: p<0.0001). The majority of the bovine strain-specific genes (∼ 85%) clustered tightly into eight genomic islands, suggesting these genes were acquired through lateral gene transfer (LGT). This bovine GBS also contained an unusually high proportion of insertion sequences (4.3% of the total genome), suggesting frequent genomic rearrangement. Comparison to other mastitis-causing species of bacteria provided strong evidence for two cases of interspecies LGT within the shared bovine environment: bovine S. agalactiae with Streptococcus uberis (nisin U operon) and Streptococcus dysgalactiae subsp. dysgalactiae (lactose operon). We also found evidence for LGT, involving the salivaricin operon, between the bovine S. agalactiae strain and either Streptococcus pyogenes or Streptococcus salivarius. Our findings provide insight into mechanisms facilitating environmental adaptation and acquisition of potential virulence factors, while highlighting both the key role LGT has played in the recent evolution of the bovine S. agalactiae strain, and the importance of LGT among pathogens within a shared environment.
Copyright © 2011 Elsevier B.V. All rights reserved.
Figures


References
-
- Baker C. Group B streptococcal infections. In: Stevens D, Kaplan E, editors. Streptococcal Infections Clinical Aspects, Microbiology, and Molecular Pathogenesis. Oxford Univeristy Press; New York: 2000. pp. 222–237.
-
- Balter S, Whitney C, Schuchat A. Epidemiology of group B streptococcal infections. In: Fischetti V, Novick R, Ferreti J, Portnoy D, Rood J, editors. Gram-Positive Pathogens. ASM Press; Washington, D.C.: 2000.
-
- Basavanna S, Khandavilli S, Yuste J, Cohen JM, Hosie AH, Webb AJ, Thomas GH, Brown JS. Screening of Streptococcus pneumoniae ABC transporter mutants demonstrates that LivJHMGF, a branched-chain amino acid ABC transporter, is necessary for disease pathogenesis. Infect Immun. 2009;77:3412–3423. - PMC - PubMed
-
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Stat Methodol. 1995;57:289–300.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

