Rigidification of the autolysis loop enhances Na(+) binding to thrombin
- PMID: 21536369
- PMCID: PMC3150630
- DOI: 10.1016/j.bpc.2011.04.003
Rigidification of the autolysis loop enhances Na(+) binding to thrombin
Abstract
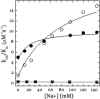
Binding of Na(+) to thrombin ensures high activity toward physiological substrates and optimizes the procoagulant and prothrombotic roles of the enzyme in vivo. Under physiological conditions of pH and temperature, the binding affinity of Na(+) is weak due to large heat capacity and enthalpy changes associated with binding, and the K(d)=80 mM ensures only 64% saturation of the site at the concentration of Na(+) in the blood (140 mM). Residues controlling Na(+) binding and activation have been identified. Yet, attempts to improve the interaction of Na(+) with thrombin and possibly increase catalytic activity under physiological conditions have so far been unsuccessful. Here we report how replacement of the flexible autolysis loop of human thrombin with the homologous rigid domain of the murine enzyme results in a drastic (up to 10-fold) increase in Na(+) affinity and a significant improvement in the catalytic activity of the enzyme. Rigidification of the autolysis loop abolishes the heat capacity change associated with Na(+) binding observed in the wild-type and also increases the stability of thrombin. These findings have general relevance to protein engineering studies of clotting proteases and trypsin-like enzymes.
Copyright © 2011 Elsevier B.V. All rights reserved.
Figures





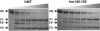

Similar articles
-
Redesigning allosteric activation in an enzyme.Proc Natl Acad Sci U S A. 2011 Mar 29;108(13):5221-5. doi: 10.1073/pnas.1018860108. Epub 2011 Feb 22. Proc Natl Acad Sci U S A. 2011. PMID: 21368156 Free PMC article.
-
Molecular dissection of Na+ binding to thrombin.J Biol Chem. 2004 Jul 23;279(30):31842-53. doi: 10.1074/jbc.M401756200. Epub 2004 May 19. J Biol Chem. 2004. PMID: 15152000
-
The Na+ binding site of thrombin.J Biol Chem. 1995 Sep 22;270(38):22089-92. doi: 10.1074/jbc.270.38.22089. J Biol Chem. 1995. PMID: 7673182
-
Thrombin allostery.Phys Chem Chem Phys. 2007 Mar 21;9(11):1291-306. doi: 10.1039/b616819a. Epub 2007 Jan 12. Phys Chem Chem Phys. 2007. PMID: 17347701 Review.
-
How Na+ activates thrombin--a review of the functional and structural data.Biol Chem. 2008 Aug;389(8):1025-35. doi: 10.1515/BC.2008.113. Biol Chem. 2008. PMID: 18979627 Free PMC article. Review.
Cited by
-
Loop Electrostatics Asymmetry Modulates the Preexisting Conformational Equilibrium in Thrombin.Biochemistry. 2016 Jul 19;55(28):3984-94. doi: 10.1021/acs.biochem.6b00385. Epub 2016 Jul 6. Biochemistry. 2016. PMID: 27347732 Free PMC article.
-
Molecular dynamics simulations of aptamer-binding reveal generalized allostery in thrombin.J Biomol Struct Dyn. 2017 Nov;35(15):3354-3369. doi: 10.1080/07391102.2016.1254682. Epub 2016 Nov 29. J Biomol Struct Dyn. 2017. PMID: 27794633 Free PMC article.
-
Isothermal titration calorimetry of ion-coupled membrane transporters.Methods. 2015 Apr;76:171-182. doi: 10.1016/j.ymeth.2015.01.012. Epub 2015 Feb 9. Methods. 2015. PMID: 25676707 Free PMC article.
-
Dabigatran and Argatroban Diametrically Modulate Thrombin Exosite Function.PLoS One. 2016 Jun 15;11(6):e0157471. doi: 10.1371/journal.pone.0157471. eCollection 2016. PLoS One. 2016. PMID: 27305147 Free PMC article.
-
Binding thermodynamics of a glutamate transporter homolog.Nat Struct Mol Biol. 2013 May;20(5):634-40. doi: 10.1038/nsmb.2548. Epub 2013 Apr 7. Nat Struct Mol Biol. 2013. PMID: 23563139 Free PMC article.
References
-
- Wyman J, Gill SJ. Binding and Linkage. University Science Books; Mill Valley, CA: 1990.
-
- Davie EW, Kulman JD. An overview of the structure and function of thrombin. Semin Thromb Hemost. 2006;32(Suppl 1):3–15. - PubMed
-
- Bode W. Structure and interaction modes of thrombin. Blood Cells Mol Dis. 2006;36:122–130. - PubMed
-
- Di Cera E, Guinto ER, Vindigni A, Dang QD, Ayala YM, Wuyi M, Tulinsky A. The Na+ binding site of thrombin. J Biol Chem. 1995;270:22089–22092. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

