The TFIIF-like Rpc37/53 dimer lies at the center of a protein network to connect TFIIIC, Bdp1, and the RNA polymerase III active center
- PMID: 21536656
- PMCID: PMC3133384
- DOI: 10.1128/MCB.05151-11
The TFIIF-like Rpc37/53 dimer lies at the center of a protein network to connect TFIIIC, Bdp1, and the RNA polymerase III active center
Abstract
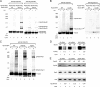
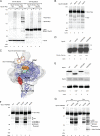
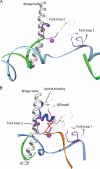
Eukaryotic RNA polymerase III (Pol III) relies on a transcription factor TFIIF-like Rpc37/53 subcomplex for promoter opening, elongation, termination, and reinitiation. By incorporating the photoreactive amino acid p-benzoyl-L-phenylalanine (BPA) into Rpc37, Rpc53, and the Rpc2 subunit of Pol III, we mapped protein-protein interactions, revealing the position of Rpc37/53 within the Pol III preinitiation complex (PIC). BPA photo-cross-linking was combined with site-directed hydroxyl radical probing to localize the Rpc37/53 dimerization module on the lobe/external 2 domains of Rpc2, in similarity to the binding of TFIIF on Pol II. N terminal to the dimerization domain, Rpc53 binds the Pol III-specific subunits Rpc82 and Rpc34, the Pol III stalk, and the assembly factor TFIIIC, essential for PIC formation. The C-terminal domain of Rpc37 interacts extensively with Rpc2 and Rpc34 and contains binding sites for initiation factor Bdp1. We also located the C-terminal domain of Rpc37 within the Pol III active center in the ternary elongation complex, where it likely functions in accurate termination. Our work explains how the Rpc37/53 dimer is anchored on the Pol III core and acts as a hub to integrate a protein network for initiation and termination.
Figures



References
-
- Brachmann C. B., et al. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115–132 - PubMed
-
- Braun B. R., Bartholomew B., Kassavetis G. A., Geiduschek E. P. 1992. Topography of transcription factor complexes on the Saccharomyces cerevisiae 5 S RNA gene. J. Mol. Biol. 228:1063–1077 - PubMed
-
- Brow D. A., Guthrie C. 1990. Transcription of a yeast U6 snRNA gene requires a polymerase III promoter element in a novel position. Genes Dev. 4:1345–1356 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
