Identification of potential conserved RNA secondary structure throughout influenza A coding regions
- PMID: 21536710
- PMCID: PMC3096049
- DOI: 10.1261/rna.2619511
Identification of potential conserved RNA secondary structure throughout influenza A coding regions
Abstract
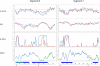
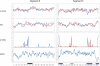
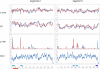
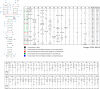
Influenza A is a negative sense RNA virus of significant public health concern. While much is understood about the life cycle of the virus, knowledge of RNA secondary structure in influenza A virus is sparse. Predictions of RNA secondary structure can focus experimental efforts. The present study analyzes coding regions of the eight viral genome segments in both the (+) and (-) sense RNA for conserved secondary structure. The predictions are based on identifying regions of unusual thermodynamic stabilities and are correlated with studies of suppression of synonymous codon usage (SSCU). The results indicate that secondary structure is favored in the (+) sense influenza RNA. Twenty regions with putative conserved RNA structure have been identified, including two previously described structured regions. Of these predictions, eight have high thermodynamic stability and SSCU, with five of these corresponding to current annotations (e.g., splice sites), while the remaining 12 are predicted by the thermodynamics alone. Secondary structures with high conservation of base-pairing are proposed within the five regions having known function. A combination of thermodynamics, amino acid and nucleotide sequence comparisons along with SSCU was essential for revealing potential secondary structures.
Figures







Similar articles
-
Identification of conserved RNA secondary structures at influenza B and C splice sites reveals similarities and differences between influenza A, B, and C.BMC Res Notes. 2014 Jan 9;7:22. doi: 10.1186/1756-0500-7-22. BMC Res Notes. 2014. PMID: 24405943 Free PMC article.
-
Codon conservation in the influenza A virus genome defines RNA packaging signals.Nucleic Acids Res. 2007;35(6):1897-907. doi: 10.1093/nar/gkm087. Epub 2007 Mar 1. Nucleic Acids Res. 2007. PMID: 17332012 Free PMC article.
-
[Influence of Synonymous Codon Bias on the RNA Secondary Structure in Influenza-A Viruses].Bing Du Xue Bao. 2016 Nov;32(6):773-81. Bing Du Xue Bao. 2016. PMID: 30004651 Chinese.
-
[Classification and genome structure of influenza virus].Nihon Rinsho. 2003 Nov;61(11):1886-91. Nihon Rinsho. 2003. PMID: 14619426 Review. Japanese.
-
RNA structure interactions and ribonucleoprotein processes of the influenza A virus.Brief Funct Genomics. 2018 Nov 26;17(6):402-414. doi: 10.1093/bfgp/elx028. Brief Funct Genomics. 2018. PMID: 29040388 Free PMC article. Review.
Cited by
-
RNase L targets distinct sites in influenza A virus RNAs.J Virol. 2015 Mar;89(5):2764-76. doi: 10.1128/JVI.02953-14. Epub 2014 Dec 24. J Virol. 2015. PMID: 25540362 Free PMC article.
-
Mutations Designed by Ensemble Defect to Misfold Conserved RNA Structures of Influenza A Segments 7 and 8 Affect Splicing and Attenuate Viral Replication in Cell Culture.PLoS One. 2016 Jun 7;11(6):e0156906. doi: 10.1371/journal.pone.0156906. eCollection 2016. PLoS One. 2016. PMID: 27272307 Free PMC article.
-
Influenza A virus PB1-F2 protein expression is regulated in a strain-specific manner by sequences located downstream of the PB1-F2 initiation codon.J Virol. 2013 Oct;87(19):10687-99. doi: 10.1128/JVI.01520-13. Epub 2013 Jul 24. J Virol. 2013. PMID: 23885074 Free PMC article.
-
The 3' splice site of influenza A segment 7 mRNA can exist in two conformations: a pseudoknot and a hairpin.PLoS One. 2012;7(6):e38323. doi: 10.1371/journal.pone.0038323. Epub 2012 Jun 7. PLoS One. 2012. PMID: 22685560 Free PMC article.
-
In vivo analysis of influenza A mRNA secondary structures identifies critical regulatory motifs.Nucleic Acids Res. 2019 Jul 26;47(13):7003-7017. doi: 10.1093/nar/gkz318. Nucleic Acids Res. 2019. PMID: 31053845 Free PMC article.
References
-
- Basler CF, Reid AH, Dybing JK, Janczewski TA, Fanning TG, Zheng H, Salvatore M, Perdue ML, Swayne DE, García-Sastre A 2001. Sequence of the 1918 pandemic influenza virus nonstructural gene (NS) segment and characterization of recombinant viruses bearing the 1918 NS genes. Proc Natl Acad Sci 98: 2746–2751 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
