A novel function of the proneural factor Ascl1 in progenitor proliferation identified by genome-wide characterization of its targets
- PMID: 21536733
- PMCID: PMC3084027
- DOI: 10.1101/gad.627811
A novel function of the proneural factor Ascl1 in progenitor proliferation identified by genome-wide characterization of its targets
Abstract
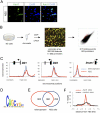
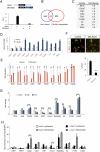
Proneural genes such as Ascl1 are known to promote cell cycle exit and neuronal differentiation when expressed in neural progenitor cells. The mechanisms by which proneural genes activate neurogenesis--and, in particular, the genes that they regulate--however, are mostly unknown. We performed a genome-wide characterization of the transcriptional targets of Ascl1 in the embryonic brain and in neural stem cell cultures by location analysis and expression profiling of embryos overexpressing or mutant for Ascl1. The wide range of molecular and cellular functions represented among these targets suggests that Ascl1 directly controls the specification of neural progenitors as well as the later steps of neuronal differentiation and neurite outgrowth. Surprisingly, Ascl1 also regulates the expression of a large number of genes involved in cell cycle progression, including canonical cell cycle regulators and oncogenic transcription factors. Mutational analysis in the embryonic brain and manipulation of Ascl1 activity in neural stem cell cultures revealed that Ascl1 is indeed required for normal proliferation of neural progenitors. This study identified a novel and unexpected activity of the proneural gene Ascl1, and revealed a direct molecular link between the phase of expansion of neural progenitors and the subsequent phases of cell cycle exit and neuronal differentiation.
Figures
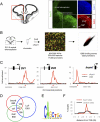
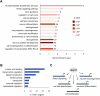
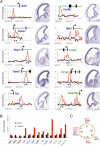
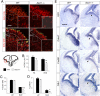

References
-
- Alonso MM, Alemany R, Fueyo J, Gomez-Manzano C 2008. E2F1 in gliomas: a paradigm of oncogene addiction. Cancer Lett 263: 157–163 - PubMed
-
- Anderson SA, Qiu M, Bulfone A, Eisenstat DD, Meneses J, Pedersen R, Rubenstein JL 1997. Mutations of the homeobox genes Dlx-1 and Dlx-2 disrupt the striatal subventricular zone and differentiation of late born striatal neurons. Neuron 19: 27–37 - PubMed
-
- Ball DW 2004. Achaete-scute homolog-1 and Notch in lung neuroendocrine development and cancer. Cancer Lett 204: 159–169 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
