RNA 3' processing functions of Arabidopsis FCA and FPA limit intergenic transcription
- PMID: 21536901
- PMCID: PMC3100917
- DOI: 10.1073/pnas.1105334108
RNA 3' processing functions of Arabidopsis FCA and FPA limit intergenic transcription
Abstract
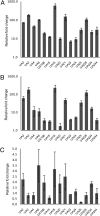
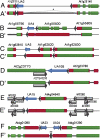
The RNA-binding proteins FCA and FPA were identified based on their repression of the flowering time regulator FLC but have since been shown to have widespread roles in the Arabidopsis thaliana genome. Here, we use whole-genome tiling arrays to show that a wide spectrum of genes and transposable elements are misexpressed in the fca-9 fpa-7 (fcafpa) double mutant at two stages of seedling development. There was a significant bias for misregulated genomic segments mapping to the 3' region of genes. In addition, the double mutant misexpressed a large number of previously unannotated genomic segments corresponding to intergenic regions. We characterized a subset of these misexpressed unannotated segments and established that they resulted from extensive transcriptional read-through, use of downstream polyadenylation sites, and alternative splicing. In some cases, the transcriptional read-through significantly reduced expression of the associated genes. FCA/FPA-dependent changes in DNA methylation were found at several loci, supporting previous associations of FCA/FPA function with chromatin modifications. Our data suggest that FCA and FPA play important roles in the A. thaliana genome in RNA 3' processing and transcription termination, thus limiting intergenic transcription.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

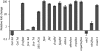

References
-
- Koornneef M, Hanhart CJ, van der Veen JH. A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol Gen Genet. 1991;229:57–66. - PubMed
-
- Macknight R, et al. FCA, a gene controlling flowering time in Arabidopsis, encodes a protein containing RNA-binding domains. Cell. 1997;89:737–745. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

