Interaction between the RNA binding domains of Ser-Arg splicing factor 1 and U1-70K snRNP protein determines early spliceosome assembly
- PMID: 21536904
- PMCID: PMC3100968
- DOI: 10.1073/pnas.1017700108
Interaction between the RNA binding domains of Ser-Arg splicing factor 1 and U1-70K snRNP protein determines early spliceosome assembly
Abstract
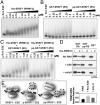
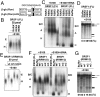
It has been widely accepted that the early spliceosome assembly begins with U1 small nuclear ribonucleoprotein (U1 snRNP) binding to the 5' splice site (5'SS), which is assisted by the Ser/Arg (SR)-rich proteins in mammalian cells. In this process, the RS domain of SR proteins is thought to directly interact with the RS motif of U1-70K, which is subject to regulation by RS domain phosphorylation. Here we report that the early spliceosome assembly event is mediated by the RNA recognition domains (RRM) of serine/arginine-rich splicing factor 1 (SRSF1), which bridges the RRM of U1-70K to pre-mRNA by using the surface opposite to the RNA binding site. Specific mutation in the RRM of SRSF1 that disrupted the RRM-RRM interaction also inhibits the formation of spliceosomal E complex and splicing. We further demonstrate that the hypo-phosphorylated RS domain of SRSF1 interacts with its own RRM, thus competing with U1-70K binding, whereas the hyper-phosphorylated RS domain permits the formation of a ternary complex containing ESE, an SR protein, and U1 snRNP. Therefore, phosphorylation of the RS domain in SRSF1 appears to induce a key molecular switch from intra- to intermolecular interactions, suggesting a plausible mechanism for the documented requirement for the phosphorylation/dephosphorylation cycle during pre-mRNA splicing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

