Effect of glycans and the glycophosphatidylinositol anchor on strain dependent conformations of scrapie prion protein: improved purifications and infrared spectra
- PMID: 21539311
- PMCID: PMC3101284
- DOI: 10.1021/bi2003907
Effect of glycans and the glycophosphatidylinositol anchor on strain dependent conformations of scrapie prion protein: improved purifications and infrared spectra
Abstract
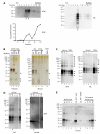
Mammalian prion diseases involve conversion of normal prion protein, PrP(C), to a pathological aggregated state (PrP(res)). The three-dimensional structure of PrP(res) is not known, but infrared (IR) spectroscopy has indicated high, strain-dependent β-sheet content. PrP(res) molecules usually contain a glycophosphatidylinositol (GPI) anchor and large Asn-linked glycans, which can also vary with strain. Using IR spectroscopy, we tested the conformational effects of these post-translational modifications by comparing wild-type PrP(res) with GPI- and glycan-deficient PrP(res) produced in GPI-anchorless PrP transgenic mice. These analyses required the development of substantially improved purification protocols. Spectra of both types of PrP(res) revealed conformational differences between the 22L, ME7, and Chandler (RML) murine scrapie strains, most notably in bands attributed to β-sheets. These PrP(res) spectra were also distinct from those of the hamster 263K scrapie strain. Spectra of wild-type and anchorless 22L PrP(res) were nearly indistinguishable. With ME7 PrP(res), modest differences between the wild-type and anchorless spectra were detected, notably an ∼2 cm(-1) shift in an apparent β-sheet band. Collectively, the data provide evidence that the glycans and anchor do not grossly affect the strain-specific secondary structures of PrP(res), at least relative to the differences observed between strains, but can subtly affect turns and certain β-sheet components. Recently reported H-D exchange analyses of anchorless PrP(res) preparations strongly suggested the presence of strain-dependent, solvent-inaccessible β-core structures throughout most of the C-terminal half of PrP(res) molecules, with no remaining α-helix. Our IR data provide evidence that similar core structures also comprise wild-type PrP(res).
Figures


References
-
- Riek R, Hornemann S, Wider G, Glockshuber R, Wuthrich K. NMR characterization of the full-length recombinant murine prion protein, mPrP(23-231) FEBS Lett. 1997;413:282–288. - PubMed
-
- Riek R, Hornemann S, Wider G, Billeter M, Glockshuber R, Wuthrich K. NMR structure of the mouse prion protein domain PrP(121-231) Nature. 1996;382:180–182. - PubMed
-
- Caughey BW, Dong A, Bhat KS, Ernst D, Hayes SF, Caughey WS. Secondary structure analysis of the scrapie-associated protein PrP 27-30 in water by infrared spectroscopy. Biochemistry. 1991;30:7672–7680. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

