A new method for isolation of interstitial fluid from human solid tumors applied to proteomic analysis of ovarian carcinoma tissue
- PMID: 21541282
- PMCID: PMC3082557
- DOI: 10.1371/journal.pone.0019217
A new method for isolation of interstitial fluid from human solid tumors applied to proteomic analysis of ovarian carcinoma tissue
Abstract
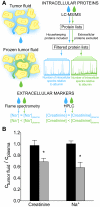
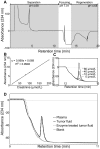
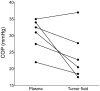
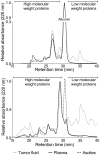
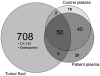
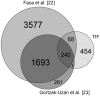
Major efforts have been invested in the identification of cancer biomarkers in plasma, but the extraordinary dynamic range in protein composition, and the dilution of disease specific proteins make discovery in plasma challenging. Focus is shifting towards using proximal fluids for biomarker discovery, but methods to verify the isolated sample's origin are missing. We therefore aimed to develop a technique to search for potential candidate proteins in the proximal proteome, i.e. in the tumor interstitial fluid, since the biomarkers are likely to be excreted or derive from the tumor microenvironment. Since tumor interstitial fluid is not readily accessible, we applied a centrifugation method developed in experimental animals and asked whether interstitial fluid from human tissue could be isolated, using ovarian carcinoma as a model. Exposure of extirpated tissue to 106 g enabled tumor fluid isolation. The fluid was verified as interstitial by an isolated fluid:plasma ratio not significantly different from 1.0 for both creatinine and Na(+), two substances predominantly present in interstitial fluid. The isolated fluid had a colloid osmotic pressure 79% of that in plasma, suggesting that there was some sieving of proteins at the capillary wall. Using a proteomic approach we detected 769 proteins in the isolated interstitial fluid, sixfold higher than in patient plasma. We conclude that the isolated fluid represents undiluted interstitial fluid and thus a subproteome with high concentration of locally secreted proteins that may be detected in plasma for diagnostic, therapeutic and prognostic monitoring by targeted methods.
Conflict of interest statement
Figures
Similar articles
-
Increased WD-repeat containing protein 1 in interstitial fluid from ovarian carcinomas shown by comparative proteomic analysis of malignant and healthy gynecological tissue.Biochim Biophys Acta. 2013 Nov;1834(11):2347-59. doi: 10.1016/j.bbapap.2013.05.011. Epub 2013 May 21. Biochim Biophys Acta. 2013. PMID: 23707566
-
Isolation of interstitial fluid from rat mammary tumors by a centrifugation method.Am J Physiol Heart Circ Physiol. 2003 Jan;284(1):H416-24. doi: 10.1152/ajpheart.00327.2002. Epub 2002 Sep 12. Am J Physiol Heart Circ Physiol. 2003. PMID: 12388326
-
Isolation of rat trachea interstitial fluid and demonstration of local cytokine production in lipopolysaccharide-induced systemic inflammation.J Appl Physiol (1985). 2008 Mar;104(3):809-20. doi: 10.1152/japplphysiol.00846.2007. Epub 2008 Jan 10. J Appl Physiol (1985). 2008. PMID: 18187613
-
Tumor interstitial fluid - a treasure trove of cancer biomarkers.Biochim Biophys Acta. 2013 Nov;1834(11):2259-70. doi: 10.1016/j.bbapap.2013.01.013. Epub 2013 Feb 14. Biochim Biophys Acta. 2013. PMID: 23416532 Review.
-
Interstitial fluid-a reflection of the tumor cell microenvironment and secretome.Biochim Biophys Acta. 2013 Nov;1834(11):2336-46. doi: 10.1016/j.bbapap.2013.01.028. Epub 2013 Jan 31. Biochim Biophys Acta. 2013. PMID: 23376185 Review.
Cited by
-
Interstitial Fluid in Gynecologic Tumors and Its Possible Application in the Clinical Practice.Int J Mol Sci. 2018 Dec 12;19(12):4018. doi: 10.3390/ijms19124018. Int J Mol Sci. 2018. PMID: 30545144 Free PMC article. Review.
-
Quantification of microenvironmental metabolites in murine cancers reveals determinants of tumor nutrient availability.Elife. 2019 Apr 16;8:e44235. doi: 10.7554/eLife.44235. Elife. 2019. PMID: 30990168 Free PMC article.
-
The Angiogenic Secretome in VEGF overexpressing Breast Cancer Xenografts.Sci Rep. 2016 Dec 20;6:39460. doi: 10.1038/srep39460. Sci Rep. 2016. PMID: 27995973 Free PMC article.
-
Ionic immune suppression within the tumour microenvironment limits T cell effector function.Nature. 2016 Sep 22;537(7621):539-543. doi: 10.1038/nature19364. Epub 2016 Sep 14. Nature. 2016. PMID: 27626381 Free PMC article.
-
Isolation and Quantification of Metabolite Levels in Murine Tumor Interstitial Fluid by LC/MS.Bio Protoc. 2019 Nov 20;9(22):e3427. doi: 10.21769/BioProtoc.3427. eCollection 2019 Nov 20. Bio Protoc. 2019. PMID: 33654924 Free PMC article.
References
-
- Hanash SM, Pitteri SJ, Faca VM. Mining the plasma proteome for cancer biomarkers. Nature. 2008;452:571–579. - PubMed
-
- Kischel P, Waltregny D, Castronovo V. Identification of accessible human cancer biomarkers using ex vivo chemical proteomic strategies. Expert Rev Proteomics. 2007;4:727–739. - PubMed
-
- Cadron I, Van Gorp T, Timmerman D, Amant F, Waelkens E, et al. Application of proteomics in ovarian cancer: which sample should be used? Gynecol Oncol. 2009;115:497–503. - PubMed
-
- Williams TI, Toups KL, Saggese DA, Kalli KR, Cliby WA, et al. Epithelial ovarian cancer: disease etiology, treatment, detection, and investigational gene, metabolite, and protein biomarkers. J Proteome Res. 2007;6:2936–2962. - PubMed
-
- Simpson RJ, Bernhard OK, Greening DW, Moritz RL. Proteomics-driven cancer biomarker discovery: looking to the future. Curr Opin Chem Biol. 2008;12:72–77. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

