Expression of a dominant negative CELF protein in vivo leads to altered muscle organization, fiber size, and subtype
- PMID: 21541285
- PMCID: PMC3082560
- DOI: 10.1371/journal.pone.0019274
Expression of a dominant negative CELF protein in vivo leads to altered muscle organization, fiber size, and subtype
Abstract
Background: CUG-BP and ETR-3-like factor (CELF) proteins regulate tissue- and developmental stage-specific alternative splicing in striated muscle. We previously demonstrated that heart muscle-specific expression of a nuclear dominant negative CELF protein in transgenic mice (MHC-CELFΔ) effectively disrupts endogenous CELF activity in the heart in vivo, resulting in impaired cardiac function. In this study, transgenic mice that express the dominant negative protein under a skeletal muscle-specific promoter (Myo-CELFΔ) were generated to investigate the role of CELF-mediated alternative splicing programs in normal skeletal muscle.
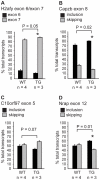
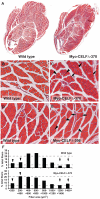
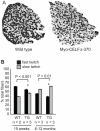
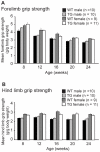
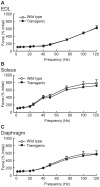
Methodology/principal findings: Myo-CELFΔ mice exhibit modest changes in CELF-mediated alternative splicing in skeletal muscle, accompanied by a reduction of endomysial and perimysial spaces, an increase in fiber size variability, and an increase in slow twitch muscle fibers. Weight gain and mean body weight, total number of muscle fibers, and overall muscle strength were not affected.
Conclusions/significance: Although these findings demonstrate that CELF activity contributes to the normal alternative splicing of a subset of muscle transcripts in vivo, the mildness of the effects in Myo-CELFΔ muscles compared to those in MHC-CELFΔ hearts suggests CELF activity may be less determinative for alternative splicing in skeletal muscle than in heart muscle. Nonetheless, even these small changes in CELF-mediated splicing regulation were sufficient to alter muscle organization and muscle fiber properties affected in myotonic dystrophy. This lends further evidence to the hypothesis that dysregulation of CELF-mediated alternative splicing programs may be responsible for the disruption of these properties during muscle pathogenesis.
Conflict of interest statement
Figures

References
-
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40(12):1413–1415. - PubMed
-
- Charlet-B N, Logan P, Singh G, Cooper T. Dynamic antagonism between ETR-3 and PTB regulates cell type-specific alternative splicing. Mol Cell. 2002;9:649–658. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

