The energy computation paradox and ab initio protein folding
- PMID: 21541343
- PMCID: PMC3081830
- DOI: 10.1371/journal.pone.0018868
The energy computation paradox and ab initio protein folding
Abstract
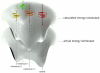
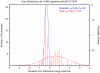
The routine prediction of three-dimensional protein structure from sequence remains a challenge in computational biochemistry. It has been intuited that calculated energies from physics-based scoring functions are able to distinguish native from nonnative folds based on previous performance with small proteins and that conformational sampling is the fundamental bottleneck to successful folding. We demonstrate that as protein size increases, errors in the computed energies become a significant problem. We show, by using error probability density functions, that physics-based scores contain significant systematic and random errors relative to accurate reference energies. These errors propagate throughout an entire protein and distort its energy landscape to such an extent that modern scoring functions should have little chance of success in finding the free energy minima of large proteins. Nonetheless, by understanding errors in physics-based score functions, they can be reduced in a post-hoc manner, improving accuracy in energy computation and fold discrimination.
Conflict of interest statement
Figures


References
-
- Anfinsen CB. Influences of 3-Dimensional Configuration on Chemical Reactivity and Stability of Proteins. Journal of Polymer Science. 1961;49:31–49.
-
- Anfinsen CB. Principles That Govern Folding of Protein Chains. Science. 1973;181:223–230. - PubMed
-
- Sohl JL, Jaswal SS, Agard DA. Unfolded conformations of alpha-lytic protease are more stable than its native state. Nature. 1998;395:817–819. - PubMed
-
- Baker D, Agard DA. Kinetics Versus Thermodynamics in Protein-Folding. Biochemistry. 1994;33:7505–7509. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

