Lack of global meiotic sex chromosome inactivation, and paucity of tissue-specific gene expression on the Drosophila X chromosome
- PMID: 21542906
- PMCID: PMC3104377
- DOI: 10.1186/1741-7007-9-29
Lack of global meiotic sex chromosome inactivation, and paucity of tissue-specific gene expression on the Drosophila X chromosome
Abstract
Background: Paucity of male-biased genes on the Drosophila X chromosome is a well-established phenomenon, thought to be specifically linked to the role of these genes in reproduction and/or their expression in the meiotic male germline. In particular, meiotic sex chromosome inactivation (MSCI) has been widely considered a driving force behind depletion of spermatocyte-biased X-linked genes in Drosophila by analogy with mammals, even though the existence of global MCSI in Drosophila has not been proven.
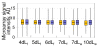
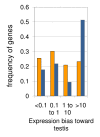
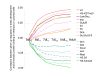
Results: Microarray-based study and qRT-PCR analyses show that the dynamics of gene expression during testis development are very similar between X-linked and autosomal genes, with both showing transcriptional activation concomitant with meiosis. However, the genes showing at least ten-fold expression bias toward testis are significantly underrepresented on the X chromosome. Intriguingly, the genes with similar expression bias toward tissues other than testis, even those not apparently associated with reproduction, are also strongly underrepresented on the X. Bioinformatics analysis shows that while tissue-specific genes often bind silencing-associated factors in embryonic and cultured cells, this trend is less prominent for the X-linked genes.
Conclusions: Our data show that the global meiotic inactivation of the X chromosome does not occur in Drosophila. Paucity of testis-biased genes on the X appears not to be linked to reproduction or germline-specific events, but rather reflects a general underrepresentation of tissue-biased genes on this chromosome. Our analyses suggest that the activation/repression switch mechanisms that probably orchestrate the highly-biased expression of tissue-specific genes are generally not efficient on the X chromosome. This effect, probably caused by dosage compensation counteracting repression of the X-linked genes, may be the cause of the exodus of highly tissue-biased genes to the autosomes.
Figures
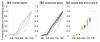

Comment in
-
Sex, sex chromosomes and gene expression.BMC Biol. 2011 May 4;9:30. doi: 10.1186/1741-7007-9-30. BMC Biol. 2011. PMID: 21542945 Free PMC article.
-
Re-analysis of the larval testis data on meiotic sex chromosome inactivation revealed evidence for tissue-specific gene expression related to the drosophila X chromosome.BMC Biol. 2012 Jun 12;10:49; author reply 50. doi: 10.1186/1741-7007-10-49. BMC Biol. 2012. PMID: 22691264 Free PMC article.
Similar articles
-
Re-analysis of the larval testis data on meiotic sex chromosome inactivation revealed evidence for tissue-specific gene expression related to the drosophila X chromosome.BMC Biol. 2012 Jun 12;10:49; author reply 50. doi: 10.1186/1741-7007-10-49. BMC Biol. 2012. PMID: 22691264 Free PMC article.
-
Stage-specific expression profiling of Drosophila spermatogenesis suggests that meiotic sex chromosome inactivation drives genomic relocation of testis-expressed genes.PLoS Genet. 2009 Nov;5(11):e1000731. doi: 10.1371/journal.pgen.1000731. Epub 2009 Nov 20. PLoS Genet. 2009. PMID: 19936020 Free PMC article.
-
Segmental dataset and whole body expression data do not support the hypothesis that non-random movement is an intrinsic property of Drosophila retrogenes.BMC Evol Biol. 2012 Sep 5;12:169. doi: 10.1186/1471-2148-12-169. BMC Evol Biol. 2012. PMID: 22950647 Free PMC article.
-
Meiotic sex chromosome inactivation in Drosophila.J Genomics. 2014 Jun 1;2:104-17. doi: 10.7150/jgen.8178. eCollection 2014. J Genomics. 2014. PMID: 25057326 Free PMC article. Review.
-
Mammalian Sex Chromosome Structure, Gene Content, and Function in Male Fertility.Annu Rev Anim Biosci. 2019 Feb 15;7:103-124. doi: 10.1146/annurev-animal-020518-115332. Epub 2018 Nov 9. Annu Rev Anim Biosci. 2019. PMID: 30412673 Review.
Cited by
-
Faster-X evolution of gene expression in Drosophila.PLoS Genet. 2012;8(10):e1003013. doi: 10.1371/journal.pgen.1003013. Epub 2012 Oct 11. PLoS Genet. 2012. PMID: 23071459 Free PMC article.
-
Dosage Compensation and the Distribution of Sex-Biased Gene Expression in Drosophila: Considerations and Genomic Constraints.J Mol Evol. 2016 May;82(4-5):199-206. doi: 10.1007/s00239-016-9735-y. Epub 2016 Apr 8. J Mol Evol. 2016. PMID: 27059220 Free PMC article.
-
Sex and speciation: Drosophila reproductive tract proteins- twenty five years later.Int J Evol Biol. 2012;2012:191495. doi: 10.1155/2012/191495. Epub 2012 Oct 17. Int J Evol Biol. 2012. PMID: 23119225 Free PMC article.
-
Rapid Functional and Sequence Differentiation of a Tandemly Repeated Species-Specific Multigene Family in Drosophila.Mol Biol Evol. 2017 Jan;34(1):51-65. doi: 10.1093/molbev/msw212. Epub 2016 Oct 3. Mol Biol Evol. 2017. PMID: 27702774 Free PMC article.
-
Drosophila Pif1A is essential for spermatogenesis and is the homolog of human CCDC157, a gene associated with idiopathic NOA.Cell Death Dis. 2019 Feb 11;10(2):125. doi: 10.1038/s41419-019-1398-3. Cell Death Dis. 2019. PMID: 30741974 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

