Chemical genomics profiling of environmental chemical modulation of human nuclear receptors
- PMID: 21543282
- PMCID: PMC3237348
- DOI: 10.1289/ehp.1002952
Chemical genomics profiling of environmental chemical modulation of human nuclear receptors
Abstract
Background: The large and increasing number of chemicals released into the environment demands more efficient and cost-effective approaches for assessing environmental chemical toxicity. The U.S. Tox21 program has responded to this challenge by proposing alternative strategies for toxicity testing, among which the quantitative high-throughput screening (qHTS) paradigm has been adopted as the primary tool for generating data from screening large chemical libraries using a wide spectrum of assays.
Objectives: The goal of this study was to develop methods to evaluate the data generated from these assays to guide future assay selection and prioritization for the Tox21 program.
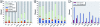
Methods: We examined the data from the Tox21 pilot-phase collection of approximately 3,000 environmental chemicals profiled in qHTS format against a panel of 10 human nuclear receptors (AR, ERα, FXR, GR, LXRβ, PPARγ, PPARδ, RXRα, TRβ, and VDR) for reproducibility, concordance of biological activity profiles with sequence homology of the receptor ligand binding domains, and structure-activity relationships.
Results: We determined the assays to be appropriate in terms of biological relevance. We found better concordance for replicate compounds for the agonist-mode than for the antagonist-mode assays, likely due to interference of cytotoxicity in the latter assays. This exercise also enabled us to formulate data-driven strategies for discriminating true signals from artifacts, and to prioritize assays based on data quality.
Conclusions: The results demonstrate the feasibility of qHTS to identify the potential for environmentally relevant chemicals to interact with key toxicity pathways related to human disease induction.
Conflict of interest statement
The views expressed in this article are those of the authors and do not necessarily reflect the views or policies of the U.S. Environmental Protection Agency. Mention of trade names or commercial products does not constitute endorsement or recommendation for use.
The authors declare they have no actual or potential competing financial interests.
Figures



References
-
- Brzozowski AM, Pike AC, Dauter Z, Hubbard RE, Bonn T, Engstrom O, et al. Molecular basis of agonism and antagonism in the oestrogen receptor. Nature. 1997;389(6652):753–758. - PubMed
-
- Butler EG, Tanaka T, Ichida T, Maruyama H, Leber AP, Williams GM. Induction of hepatic peroxisome proliferation in mice by lactofen, a diphenyl ether herbicide. Toxicol Appl Pharmacol. 1988;93(1):72–80. - PubMed
-
- Cleator SJ, Ahamed E, Coombes RC, Palmieri C. A 2009 update on the treatment of patients with hormone receptor-positive breast cancer. Clin Breast Cancer. 2009;9(suppl 1):S6–S17. - PubMed
-
- Damstra T, Barlow S, Bergman A, Kavlock R, Van Der Kraak G, eds. Geneva: World Health Organization; 2002. International Programme on Chemical Safety Global Assessment: The State-of-the-Science of Endocrine Disruptors.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

