Identification of a novel neuropathogenic Theiler's murine encephalomyelitis virus
- PMID: 21543488
- PMCID: PMC3126553
- DOI: 10.1128/JVI.00274-11
Identification of a novel neuropathogenic Theiler's murine encephalomyelitis virus
Abstract
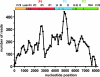
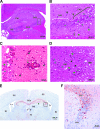
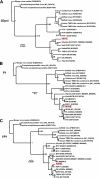
Theiler's murine encephalitis viruses (TMEV) are divided into two subgroups based on their neurovirulence. Persistent strains resemble Theiler's original viruses (referred to as the TO subgroup), which largely induce a subclinical polioencephalomyelitis during the acute phase of the disease and can persist in the spinal cord of susceptible animals, inducing a chronic demyelinating disease. In contrast, members of the neurovirulent subgroup cause an acute encephalitis characterized by the rapid onset of paralysis and death within days following intracranial inoculation. We report herein the characterization of a novel neurovirulent strain of TMEV, identified using pyrosequencing technology and referred to as NIHE. Complete coverage of the NIHE viral genome was obtained, and it shares <90% nucleotide sequence identity to known TMEV strains irrespective of subgroup, with the greatest sequence variability being observed in genes encoding the leader and capsid proteins. The histopathological analysis of infected brain and spinal cord demonstrate inflammatory lesions and neuronal necrosis during acute infection with no evidence of viral persistence or chronic disease. Intriguingly, genetic analysis indicates the putative expression of the L protein, considered a hallmark of strains within the persistent subgroup. Thus, the identification and characterization of a novel neurovirulent TMEV strain sharing features previously associated with both subgroups will lead to a deeper understanding of the evolution of TMEV strains and new insights into the determinants of neurovirulence.
Figures





References
-
- Aubert C., Chamorro M., Brahic M. 1987. Identification of Theiler's virus infected cells in the central nervous system of the mouse during demyelinating disease. Microb. Pathog. 3:319–326 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

