Neutralizing monoclonal antibodies against hepatitis C virus E2 protein bind discontinuous epitopes and inhibit infection at a postattachment step
- PMID: 21543495
- PMCID: PMC3126585
- DOI: 10.1128/JVI.00586-11
Neutralizing monoclonal antibodies against hepatitis C virus E2 protein bind discontinuous epitopes and inhibit infection at a postattachment step
Abstract
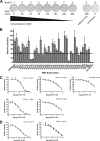
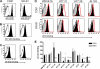
The E2 glycoprotein of hepatitis C virus (HCV) mediates viral attachment and entry into target hepatocytes and elicits neutralizing antibodies in infected patients. To characterize the structural and functional basis of HCV neutralization, we generated a novel panel of 78 monoclonal antibodies (MAbs) against E2 proteins from genotype 1a and 2a HCV strains. Using high-throughput focus-forming reduction or luciferase-based neutralization assays with chimeric infectious HCV containing structural proteins from both genotypes, we defined eight MAbs that significantly inhibited infection of the homologous HCV strain in cell culture. Two of these bound E2 proteins from strains representative of HCV genotypes 1 to 6, and one of these MAbs, H77.39, neutralized infection of strains from five of these genotypes. The three most potent neutralizing MAbs in our panel, H77.16, H77.39, and J6.36, inhibited infection at an early postattachment step. Receptor binding studies demonstrated that H77.39 inhibited binding of soluble E2 protein to both CD81 and SR-B1, J6.36 blocked attachment to SR-B1 and modestly reduced binding to CD81, and H77.16 blocked attachment to SR-B1 only. Using yeast surface display, we localized epitopes for the neutralizing MAbs on the E2 protein. Two of the strongly inhibitory MAbs, H77.16 and J6.36, showed markedly reduced binding when amino acids within hypervariable region 1 (HVR1) and at sites ∼100 to 200 residues away were changed, suggesting binding to a discontinuous epitope. Collectively, these studies help to define the structural and functional complexity of antibodies against HCV E2 protein with neutralizing potential.
Figures






References
-
- Allander T., et al. 2000. Recombinant human monoclonal antibodies against different conformational epitopes of the E2 envelope glycoprotein of hepatitis C virus that inhibit its interaction with CD81. J. Gen. Virol. 81:2451–2459 - PubMed
-
- Bartenschlager R., Frese M., Pietschmann T. 2004. Novel insights into hepatitis C virus replication and persistence. Adv. Virus Res. 63:71–180 - PubMed
-
- Bartosch B., et al. 2003. Cell entry of hepatitis C virus requires a set of co-receptors that include the CD81 tetraspanin and the SR-B1 scavenger receptor. J. Biol. Chem. 278:41624–41630 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

