The domain I-domain III linker plays an important role in the fusogenic conformational change of the alphavirus membrane fusion protein
- PMID: 21543498
- PMCID: PMC3126531
- DOI: 10.1128/JVI.00596-11
The domain I-domain III linker plays an important role in the fusogenic conformational change of the alphavirus membrane fusion protein
Abstract
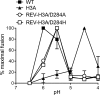
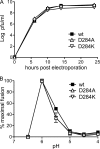
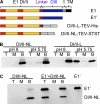
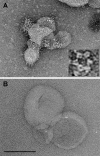
The alphavirus Semliki Forest virus (SFV) infects cells through a low-pH-dependent membrane fusion reaction mediated by the virus fusion protein E1. Acidic pH initiates a series of E1 conformational changes that culminate in membrane fusion and include dissociation of the E1/E2 heterodimer, insertion of the E1 fusion loop into the target membrane, and refolding of E1 to a stable trimeric hairpin conformation. A highly conserved histidine (H3) on the E1 protein was previously shown to promote low-pH-dependent E1 refolding. An SFV mutant with an alanine substitution at this position (H3A) has a lower pH threshold and reduced efficiency of virus fusion and E1 trimer formation than wild-type SFV. Here we addressed the mechanism by which H3 promotes E1 refolding and membrane fusion. We identified E1 mutations that rescue the H3A defect. These revertants implicated a network of interactions that connect the domain I-domain III (DI-DIII) linker region with the E1 core trimer, including H3. In support of the importance of these interactions, mutation of residues in the network resulted in more acidic pH thresholds and reduced efficiencies of membrane fusion. In vitro studies of truncated E1 proteins demonstrated that the DI-DIII linker was required for production of a stable E1 core trimer on target membranes. Together, our results suggest a critical and previously unidentified role for the DI-DIII linker region during the low-pH-dependent refolding of E1 that drives membrane fusion.
Figures



Similar articles
-
Role of conserved histidine residues in the low-pH dependence of the Semliki Forest virus fusion protein.J Virol. 2009 May;83(9):4670-7. doi: 10.1128/JVI.02646-08. Epub 2009 Feb 25. J Virol. 2009. PMID: 19244325 Free PMC article.
-
Pseudorevertants of a Semliki forest virus fusion-blocking mutation reveal a critical interchain interaction in the core trimer.J Virol. 2010 Nov;84(22):11624-33. doi: 10.1128/JVI.01625-10. Epub 2010 Sep 8. J Virol. 2010. PMID: 20826687 Free PMC article.
-
E1 mutants identify a critical region in the trimer interface of the Semliki forest virus fusion protein.J Virol. 2009 Nov;83(21):11298-306. doi: 10.1128/JVI.01147-09. Epub 2009 Aug 19. J Virol. 2009. PMID: 19692469 Free PMC article.
-
Assembly and entry mechanisms of Semliki Forest virus.Arch Virol Suppl. 1994;9:329-38. doi: 10.1007/978-3-7091-9326-6_33. Arch Virol Suppl. 1994. PMID: 8032265 Review.
-
Dealing with low pH: entry and exit of alphaviruses and flaviviruses.Trends Microbiol. 2009 Nov;17(11):514-21. doi: 10.1016/j.tim.2009.08.002. Epub 2009 Sep 30. Trends Microbiol. 2009. PMID: 19796949 Free PMC article. Review.
Cited by
-
Resveratrol Inhibits Venezuelan Equine Encephalitis Virus Infection by Interfering with the AKT/GSK Pathway.Plants (Basel). 2021 Feb 12;10(2):346. doi: 10.3390/plants10020346. Plants (Basel). 2021. PMID: 33673026 Free PMC article.
-
Evolution-Driven Attenuation of Alphaviruses Highlights Key Glycoprotein Determinants Regulating Viral Infectivity and Dissemination.Cell Rep. 2019 Jul 9;28(2):460-471.e5. doi: 10.1016/j.celrep.2019.06.022. Cell Rep. 2019. PMID: 31291581 Free PMC article.
-
Entry receptors - the gateway to alphavirus infection.J Clin Invest. 2023 Jan 17;133(2):e165307. doi: 10.1172/JCI165307. J Clin Invest. 2023. PMID: 36647825 Free PMC article. Review.
-
A key interaction between the alphavirus envelope proteins responsible for initial dimer dissociation during fusion.J Virol. 2013 Apr;87(7):3774-81. doi: 10.1128/JVI.03310-12. Epub 2013 Jan 16. J Virol. 2013. PMID: 23325694 Free PMC article.
-
Early Events in Chikungunya Virus Infection-From Virus Cell Binding to Membrane Fusion.Viruses. 2015 Jul 7;7(7):3647-74. doi: 10.3390/v7072792. Viruses. 2015. PMID: 26198242 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

