Complete genome analysis of coxsackievirus A2, A4, A5, and A10 strains isolated from hand, foot, and mouth disease patients in China revealing frequent recombination of human enterovirus A
- PMID: 21543560
- PMCID: PMC3147834
- DOI: 10.1128/JCM.00007-11
Complete genome analysis of coxsackievirus A2, A4, A5, and A10 strains isolated from hand, foot, and mouth disease patients in China revealing frequent recombination of human enterovirus A
Abstract
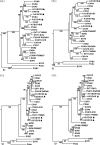
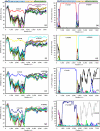
Coxsackievirus (CV) strains CVA2, CVA4, CVA5, and CVA10 were isolated from patients with hand, foot, and mouth disease during a 2009 outbreak in China. Full genome sequences for four representative strains, CVA2/SD/CHN/09 (A2SD09), CVA4/SZ/CHN/09 (A4SZ09), CVA5/SD/CHN/09 (A5SD09), and CVA10/SD/CHN/09 (A10SD09), were determined. Phylogenetic and recombination analyses of the isolates by comparison with human enterovirus A prototype strains revealed that genetic recombination occurred during cocirculation of the viruses. The A2SD09 and A4SZ09 strains were most closely related to their corresponding prototype strains in the capsid region but shared noncapsid sequences with each other. Similarly, strains A5SD09 and A10SD09 had serotype-specific homology for the capsid proteins but shared noncapsid sequences with each other. Phylogenetic analyses of the four isolates with homotypic strains showed that CVA2 strains were divided into five genotypes. The A2SD09 strain clustered with Mongolia strains isolated in 2003, forming genotype V. The A4SZ09 strain and other isolates from mainland China and Taiwan clustered with genotype III strains and are likely related to strains that circulated in Europe and Mongolia. The A5SD09 strain is closely related to other Chinese strains isolated in 2008. The A10SD09 isolate, together with other Chinese strains isolated since 2004, formed a distinct lineage that was likely imported from Japan and South Korea. This study shows that natural recombination is a frequent event in human enterovirus A evolution. More comprehensive surveillance of enteroviruses that focus not only on EV71 or CVA16 is needed for us to understand the molecular epidemiology of enteroviruses and to track recombination events which may ultimately affect the virulence of viruses during outbreaks.
Figures

References
-
- Ang L. W., et al. 2009. Epidemiology and control of hand, foot and mouth disease in Singapore, 2001-2007. Ann. Acad. Med. Singapore 38:106–112 - PubMed
-
- Blomqvist S., et al. 2010. Co-circulation of coxsackieviruses A6 and A10 in hand, foot and mouth disease outbreak in Finland. J. Clin. Virol. 48:49–54 - PubMed
-
- Caro V., Guillot S., Delpeyroux F., Crainic R. 2001. Molecular strategy for ‘serotyping' of human enteroviruses. J. Gen. Virol. 82:79–91 - PubMed
-
- Casas I., et al. 2001. Molecular characterization of human enteroviruses in clinical samples: comparison between VP2, VP1, and RNA polymerase regions using RT nested PCR assays and direct sequencing of products. J. Med. Virol. 65:138–148 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

