A cluster of coregulated genes determines TGF-beta-induced regulatory T-cell (Treg) dysfunction in NOD mice
- PMID: 21543717
- PMCID: PMC3102351
- DOI: 10.1073/pnas.1105364108
A cluster of coregulated genes determines TGF-beta-induced regulatory T-cell (Treg) dysfunction in NOD mice
Abstract
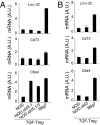
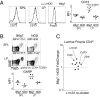
Foxp3(+) regulatory T cells (Tregs) originate in the thymus, but the Treg phenotype can also be induced in peripheral lymphoid organs or in vitro by stimulation of conventional CD4(+) T cells with IL-2 and TGF-β. There have been divergent reports on the suppressive capacity of these TGF-Treg cells. We find that TGF-Tregs derived from diabetes-prone NOD mice, although expressing normal Foxp3 levels, are uniquely defective in suppressive activity, whereas TGF-Tregs from control strains (B6g7) or ex vivo Tregs from NOD mice all function normally. Most Treg-typical transcripts were shared by NOD or B6g7 TGF-Tregs, except for a small group of differentially expressed genes, including genes relevant for suppressive activity (Lrrc32, Ctla4, and Cd73). Many of these transcripts form a coregulated cluster in a broader analysis of T-cell differentiation. The defect does not map to idd3 or idd5 regions. Whereas Treg cells from NOD mice are normal in spleen and lymph nodes, the NOD defect is observed in locations that have been tied to pathogenesis of diabetes (small intestine lamina propria and pancreatic lymph node). Thus, a genetic defect uniquely affects a specific Treg subpopulation in NOD mice, in a manner consistent with a role in determining diabetes susceptibility.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Salomon B, et al. B7/CD28 costimulation is essential for the homeostasis of the CD4+CD25+ immunoregulatory T cells that control autoimmune diabetes. Immunity. 2000;12:431–440. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

