EXORDIUM-LIKE1 promotes growth during low carbon availability in Arabidopsis
- PMID: 21543728
- PMCID: PMC3135934
- DOI: 10.1104/pp.111.177204
EXORDIUM-LIKE1 promotes growth during low carbon availability in Arabidopsis
Abstract
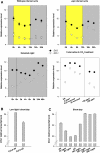
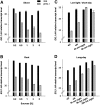
Little is known about genes that control growth and development under low carbon (C) availability. The Arabidopsis (Arabidopsis thaliana) EXORDIUM-LIKE1 (EXL1) gene (At1g35140) was identified as a brassinosteroid-regulated gene in a previous study. We show here that the EXL1 protein is required for adaptation to C- and energy-limiting growth conditions. In-depth analysis of EXL1 transcript levels under various environmental conditions indicated that EXL1 expression is controlled by the C and energy status. Sugar starvation, extended night, and anoxia stress induced EXL1 gene expression. The C status also determined EXL1 protein levels. These results suggested that EXL1 is involved in the C-starvation response. Phenotypic changes of an exl1 loss-of-function mutant became evident only under corresponding experimental conditions. The mutant showed diminished biomass production in a short-day/low-light growth regime, impaired survival during extended night, and impaired survival of anoxia stress. Basic metabolic processes and signaling pathways are presumed to be barely impaired in exl1, because the mutant showed wild-type levels of major sugars, and transcript levels of only a few genes such as QUA-QUINE STARCH were altered. Our data suggest that EXL1 is part of a regulatory pathway that controls growth and development when C and energy supply is poor.
Figures

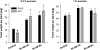
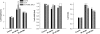


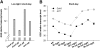
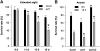
References
-
- Ahuatzi D, Riera A, Peláez R, Herrero P, Moreno F. (2007) Hxk2 regulates the phosphorylation state of Mig1 and therefore its nucleocytoplasmic distribution. J Biol Chem 282: 4485–4493 - PubMed
-
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 - PubMed
-
- Baena-González E, Rolland F, Thevelein JM, Sheen J. (2007) A central integrator of transcription networks in plant stress and energy signalling. Nature 448: 938–942 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

