The structure of the PERK kinase domain suggests the mechanism for its activation
- PMID: 21543844
- PMCID: PMC3087621
- DOI: 10.1107/S0907444911006445
The structure of the PERK kinase domain suggests the mechanism for its activation
Abstract
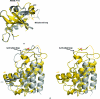
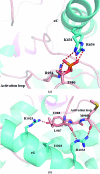
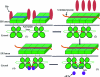
The endoplasmic reticulum (ER) unfolded protein response (UPR) is comprised of several intracellular signaling pathways that alleviate ER stress. The ER-localized transmembrane kinase PERK is one of three major ER stress transducers. Oligomerization of PERK's N-terminal ER luminal domain by ER stress promotes PERK trans-autophosphorylation of the C-terminal cytoplasmic kinase domain at multiple residues including Thr980 on the kinase activation loop. Activated PERK phosphorylates Ser51 of the α-subunit of translation initiation factor 2 (eIF2α), which inhibits initiation of protein synthesis and reduces the load of unfolded proteins entering the ER. The crystal structure of PERK's kinase domain has been determined to 2.8 Å resolution. The structure resembles the back-to-back dimer observed in the related eIF2α kinase PKR. Phosphorylation of Thr980 stabilizes both the activation loop and helix αG in the C-terminal lobe, preparing the latter for eIF2α binding. The structure suggests conservation in the mode of activation of eIF2α kinases and is consistent with a `line-up' model for PERK activation triggered by oligomerization of its luminal domain.
Figures


Similar articles
-
The luminal domain of the ER stress sensor protein PERK binds misfolded proteins and thereby triggers PERK oligomerization.J Biol Chem. 2018 Mar 16;293(11):4110-4121. doi: 10.1074/jbc.RA117.001294. Epub 2018 Jan 31. J Biol Chem. 2018. PMID: 29386355 Free PMC article.
-
The PERK/PKR-eIF2α Pathway Negatively Regulates Porcine Hemagglutinating Encephalomyelitis Virus Replication by Attenuating Global Protein Translation and Facilitating Stress Granule Formation.J Virol. 2022 Jan 12;96(1):e0169521. doi: 10.1128/JVI.01695-21. Epub 2021 Oct 13. J Virol. 2022. PMID: 34643429 Free PMC article.
-
Activation-dependent substrate recruitment by the eukaryotic translation initiation factor 2 kinase PERK.J Cell Biol. 2006 Jan 16;172(2):201-9. doi: 10.1083/jcb.200508099. J Cell Biol. 2006. PMID: 16418533 Free PMC article.
-
Endoplasmic reticulum stress in liver disease.J Hepatol. 2011 Apr;54(4):795-809. doi: 10.1016/j.jhep.2010.11.005. Epub 2010 Nov 13. J Hepatol. 2011. PMID: 21145844 Free PMC article. Review.
-
Unfolded Protein Response and PERK Kinase as a New Therapeutic Target in the Pathogenesis of Alzheimer's Disease.Curr Med Chem. 2015;22(27):3169-84. doi: 10.2174/0929867322666150818104254. Curr Med Chem. 2015. PMID: 26282939 Free PMC article. Review.
Cited by
-
A novel mechanism of PHB2-mediated mitophagy participating in the development of Parkinson's disease.Neural Regen Res. 2024 Aug 1;19(8):1828-1834. doi: 10.4103/1673-5374.389356. Epub 2023 Nov 8. Neural Regen Res. 2024. PMID: 38103250 Free PMC article.
-
Navigating the landscape of the unfolded protein response in CD8+ T cells.Front Immunol. 2024 Jul 4;15:1427859. doi: 10.3389/fimmu.2024.1427859. eCollection 2024. Front Immunol. 2024. PMID: 39026685 Free PMC article. Review.
-
Endoplasmic Reticulum Stress-Related Genes in Yellow Catfish Pelteobagrus fulvidraco: Molecular Characterization, Tissue Expression, and Expression Responses to Dietary Copper Deficiency and Excess.G3 (Bethesda). 2015 Aug 13;5(10):2091-104. doi: 10.1534/g3.115.019950. G3 (Bethesda). 2015. PMID: 26276384 Free PMC article.
-
Parallel Signaling through IRE1α and PERK Regulates Pancreatic Neuroendocrine Tumor Growth and Survival.Cancer Res. 2019 Dec 15;79(24):6190-6203. doi: 10.1158/0008-5472.CAN-19-1116. Epub 2019 Oct 31. Cancer Res. 2019. PMID: 31672843 Free PMC article.
-
Ursodeoxycholic Acid Binds PERK and Ameliorates Neurite Atrophy in a Cellular Model of GM2 Gangliosidosis.Int J Mol Sci. 2023 Apr 13;24(8):7209. doi: 10.3390/ijms24087209. Int J Mol Sci. 2023. PMID: 37108372 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

