Structure of CBM4 from Clostridium thermocellum cellulase K
- PMID: 21543854
- PMCID: PMC3087633
- DOI: 10.1107/S1744309111003307
Structure of CBM4 from Clostridium thermocellum cellulase K
Abstract
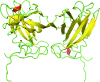
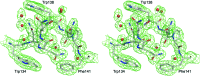
Here, a 2.0 Å resolution X-ray structure of Clostridium thermocellum cellulase K family 4 carbohydrate-binding module (CelK CBM4) is reported. The resulting structure was refined to an R factor of 0.212 and an R(free) of 0.274. Structural analysis shows that this new structure is very similar to the previously solved structure of C. thermocellum CbhA CBM4. Most importantly, these data support the previously proposed notion of an extended binding pocket using a novel tryptophan-containing loop that may be highly conserved in clostridial CBM4 proteins.
Figures

References
-
- Alahuhta, M., Xu, Q., Bomble, Y. J., Brunecky, R., Adney, W. S., Ding, S.-Y., Himmel, M. E. & Lunin, V. V. (2010). J. Mol. Biol. 402, 374–387. - PubMed
-
- Bayer, E. A., Lamed, R., White, B. A. & Flint, H. J. (2008). Chem. Rec. 8, 364–377. - PubMed
-
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources

