MicroRNA signatures differentiate melanoma subtypes
- PMID: 21543894
- PMCID: PMC3233487
- DOI: 10.4161/cc.10.11.15777
MicroRNA signatures differentiate melanoma subtypes
Abstract
Melanoma is an aggressive cancer that is highly resistance to therapies once metastasized. We studied microRNA (miRNA) expression in clinical melanoma subtypes and evaluated different miRNA signatures in the background of gain of function somatic and inherited mutations associated with melanoma. Total RNA from 42 patient derived primary melanoma cell lines and three independent normal primary melanocyte cell cultures was evaluated by miRNA array. MiRNA expression was then analyzed comparing subtypes and additional clinicopathologic criteria including somatic mutations. The prevalence and association of an inherited variant in a miRNA binding site in the 3'UTR of the KRAS oncogene, referred to as the KRAS-variant, was also evaluated. We show that seven miRNAs, miR-142-3p, miR-486, miR-214, miR-218, miR-362, miR-650 and miR-31, were significantly correlated with acral as compared to non-acral melanomas (p < 0.04). In addition, we discovered that the KRAS-variant was enriched in non-acral melanoma (25%), and that miR-137 under expression was significantly associated with melanomas with the KRAS-variant. Our findings indicate that miRNAs are differentially expressed in melanoma subtypes and that their misregulation can be impacted by inherited gene variants, supporting the hypothesis that miRNA misregulation reflects biological differences in melanoma.
Figures
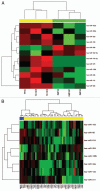
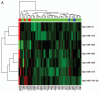


Comment in
-
MicroRNAs as molecular classifiers for cancer.Cell Cycle. 2011 Sep 1;10(17):2827-8. doi: 10.4161/cc.10.17.16574. Epub 2011 Sep 1. Cell Cycle. 2011. PMID: 21869596 Free PMC article. No abstract available.
-
New insights into the biology of melanomas using a microRNA tool-KIT.Cell Cycle. 2011 Sep 1;10(17):2828-9. Epub 2011 Sep 1. Cell Cycle. 2011. PMID: 21869597 No abstract available.
References
-
- Liu V, Mihm MC. Pathology of malignant melanoma. Surg Clin North Am. 2003;83:31–60. - PubMed
-
- Esquela-Kerscher A, Slack FJ. Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. - PubMed
-
- Babar IA, Slack FJ, Weidhaas JB. miRNA modulation of the cellular stress response. Future Oncol. 2008;4:289–298. - PubMed
-
- Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism and function. Cell. 2004;116:281–297. - PubMed
-
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous
