Variation in array size, monomer composition and expression of the macrosatellite DXZ4
- PMID: 21544201
- PMCID: PMC3081327
- DOI: 10.1371/journal.pone.0018969
Variation in array size, monomer composition and expression of the macrosatellite DXZ4
Abstract
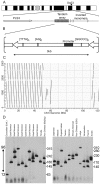
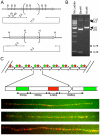
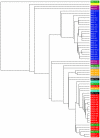
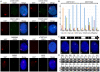
Macrosatellites are some of the most polymorphic regions of the human genome, yet many remain uncharacterized despite the association of some arrays with disease susceptibility. This study sought to explore the polymorphic nature of the X-linked macrosatellite DXZ4. Four aspects of DXZ4 were explored in detail, including tandem repeat copy number variation, array instability, monomer sequence polymorphism and array expression. DXZ4 arrays contained between 12 and 100 3.0 kb repeat units with an average array containing 57. Monomers were confirmed to be arranged in uninterrupted tandem arrays by restriction digest analysis and extended fiber FISH, and therefore DXZ4 encompasses 36-288 kb of Xq23. Transmission of DXZ4 through three generations in three families displayed a high degree of meiotic instability (8.3%), consistent with other macrosatellite arrays, further highlighting the unstable nature of these sequences in the human genome. Subcloning and sequencing of complete DXZ4 monomers identified numerous single nucleotide polymorphisms and alleles for the three microsatellite repeats located within each monomer. Pairwise comparisons of DXZ4 monomer sequences revealed that repeat units from an array are more similar to one another than those originating from different arrays. RNA fluorescence in situ hybridization revealed significant variation in DXZ4 expression both within and between cell lines. DXZ4 transcripts could be detected originiating from both the active and inactive X chromosome. Expression levels of DXZ4 varied significantly between males, but did not relate to the size of the array, nor did inheritance of the same array result in similar expression levels. Collectively, these studies provide considerable insight into the polymorphic nature of DXZ4, further highlighting the instability and variation potential of macrosatellites in the human genome.
Conflict of interest statement
Figures

References
-
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. - PubMed
-
- Giacalone J, Friedes J, Francke U. A novel GC-rich human macrosatellite VNTR in Xq24 is differentially methylated on active and inactive X chromosomes. Nat Genet. 1992;1:137–143. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

