Oncogenic KRAS-induced interleukin-8 overexpression promotes cell growth and migration and contributes to aggressive phenotypes of non-small cell lung cancer
- PMID: 21544811
- PMCID: PMC3374723
- DOI: 10.1002/ijc.26164
Oncogenic KRAS-induced interleukin-8 overexpression promotes cell growth and migration and contributes to aggressive phenotypes of non-small cell lung cancer
Abstract
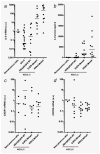
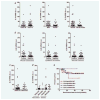
The CXC chemokine interleukin-8 (IL-8) is an angiogenic growth factor that is overexpressed in various cancers, including non-small cell lung cancer (NSCLC). Previously, IL-8 was shown as a transcriptional target of RAS signaling, raising the possibility of its role in oncogenic KRAS-driven NSCLC. Using microarray analysis, we identified IL-8 as the most downregulated gene by shRNA-mediated KRAS knockdown in NCI-H1792 NSCLC cells where IL-8 is overexpressed. NSCLC cell lines harboring KRAS or EGFR mutations overexpressed IL-8, while IL-8 levels were more prominent in KRAS mutants compared to EGFR mutants. IL-8 expression was downregulated by shRNA-mediated KRAS knockdown in KRAS mutants or by treatment with EGFR tyrosine kinase inhibitors and EGFR siRNAs in EGFR mutants. In our analysis of the relationship of IL-8 expression with clinical parameters and mutation status of KRAS or EGFR in 89 NSCLC surgical specimens, IL-8 expression was shown to be significantly higher in NSCLCs of males, smokers, and elderly patients and those with pleural involvement and KRAS mutated adenocarcinomas. In KRAS mutant cells, the MEK inhibitor markedly decreased IL-8 expression, while the p38 inhibitor increased IL-8 expression. Attenuation of IL-8 function by siRNAs or a neutralizing antibody inhibited cell proliferation and migration of KRAS mutant/IL-8 overexpressing NSCLC cells. These results indicate that activating mutations of KRAS or EGFR upregulate IL-8 expression in NSCLC; IL-8 is highly expressed in NSCLCs from males, smokers, elderly patients, NSCLCs with pleural involvement, and KRAS-mutated adenocarcinomas; and IL-8 plays a role in cell growth and migration in oncogenic KRAS-driven NSCLC.
Copyright © 2011 UICC.
Figures


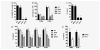
References
-
- Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer. 2003;3:11–22. - PubMed
-
- Sato M, Shames DS, Gazdar AF, Minna JD. A translational view of the molecular pathogenesis of lung cancer. J Thorac Oncol. 2007;2:327–343. - PubMed
-
- Buttitta F, Barassi F, Fresu G, Felicioni L, Chella A, Paolizzi D, Lattanzio G, Salvatore S, Camplese PP, Rosini S, Iarussi T, Mucilli F, et al. Mutational analysis of the HER2 gene in lung tumors from Caucasian patients: mutations are mainly present in adenocarcinomas with bronchioloalveolar features. Int J Cancer. 2006;119:2586–2591. - PubMed
-
- Mascaux C, Iannino N, Martin B, Paesmans M, Berghmans T, Dusart M, Haller A, Lothaire P, Meert AP, Noel S, Lafitte JJ, Sculier JP. The role of RAS oncogene in survival of patients with lung cancer: a systematic review of the literature with meta-analysis. Br J Cancer. 2005;92:131–139. - PMC - PubMed
-
- Isobe T, Herbst RS, Onn A. Current management of advanced non-small cell lung cancer: targeted therapy. Semin Oncol. 2005;32:315–328. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

