Activation of the signal transducer and activator of transcription 3 pathway up-regulates estrogen receptor-beta expression in lung adenocarcinoma cells
- PMID: 21546410
- PMCID: PMC5417244
- DOI: 10.1210/me.2010-0495
Activation of the signal transducer and activator of transcription 3 pathway up-regulates estrogen receptor-beta expression in lung adenocarcinoma cells
Abstract
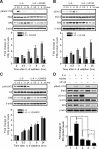
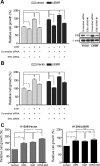
Estrogens contribute to the pathogenesis of female lung cancer and function mainly through estrogen receptor-β (ERβ). However, the way in which ERβ expression is regulated in lung cancer cells remains to be explored. We have found that signal transducer and activator of transcription 3 (Stat3) activation up-regulates ERβ expression in PC14PE6/AS2 lung cancer cells in a preliminary Affymetrix oligonucleotide array study, and we sought to confirm the findings. In this study, we show that IL-6 induced ERβ mRNA and protein expression in lung cancer cells. The induction of ERβ in response to IL-6 was abolished by Janus kinase 2 inhibitor-AG490, dominant-negative mutant of Stat3, and Stat3-targeting short interfering RNA. The luciferase reporter assay and chromatin immunoprecipitation assay confirmed that IL-6-activated Stat3 binds to the ERβ promoter. Besides the Janus kinase 2/Stat3 pathway, the MEK/Erk pathway contributes to ERβ up-regulation induced by IL-6; however, the phosphoinositide 3'-kinase/Akt pathway does not. We also found that epidermal growth factor (EGF) stimulation or L858R mutation in EGF receptor (EGFR) induced Stat3 activation as well as ERβ expression in lung cancer cells. Inhibiting Stat3 activity by pharmacological or genetic approaches reduced EGF- and L858R mutant EGFR-induced ERβ expression, indicating that Stat3 activation is required for EGFR signaling-mediated ERβ up-regulation. Silencing ERβ decreased cell proliferation in lung cancer cells that overexpress L858R mutant EGFR. In conclusion, we have identified that Stat3 activation is essential for ERβ induction by IL-6, EGF, and the presence of EGFR mutation. The findings shed light on new therapeutic targets for female lung cancer, especially for those with EGFR mutations.
Figures





Similar articles
-
Epidermal growth factor receptor cooperates with signal transducer and activator of transcription 3 to induce epithelial-mesenchymal transition in cancer cells via up-regulation of TWIST gene expression.Cancer Res. 2007 Oct 1;67(19):9066-76. doi: 10.1158/0008-5472.CAN-07-0575. Cancer Res. 2007. PMID: 17909010 Free PMC article.
-
Interleukin-6 induces proliferation in adult spinal cord-derived neural progenitors via the JAK2/STAT3 pathway with EGF-induced MAPK phosphorylation.Cell Prolif. 2008 Jun;41(3):377-92. doi: 10.1111/j.1365-2184.2008.00537.x. Cell Prolif. 2008. PMID: 18485152 Free PMC article.
-
Signal transducer and activator of transcription 3 activation up-regulates interleukin-6 autocrine production: a biochemical and genetic study of established cancer cell lines and clinical isolated human cancer cells.Mol Cancer. 2010 Dec 2;9:309. doi: 10.1186/1476-4598-9-309. Mol Cancer. 2010. PMID: 21122157 Free PMC article.
-
STAT3 as a mediator of oncogenic cellular metabolism: Pathogenic and therapeutic implications.Neoplasia. 2021 Dec;23(12):1167-1178. doi: 10.1016/j.neo.2021.10.003. Epub 2021 Oct 29. Neoplasia. 2021. PMID: 34731785 Free PMC article. Review.
-
Potential of targeting signal-transducing adaptor protein-2 in cancer therapeutic applications.Explor Target Antitumor Ther. 2024;5(2):251-259. doi: 10.37349/etat.2024.00216. Epub 2024 Mar 7. Explor Target Antitumor Ther. 2024. PMID: 38745775 Free PMC article. Review.
Cited by
-
Transcription factors and hormone receptors: Sex‑specific targets for cancer therapy (Review).Oncol Lett. 2024 Dec 6;29(2):93. doi: 10.3892/ol.2024.14839. eCollection 2025 Feb. Oncol Lett. 2024. PMID: 39691589 Free PMC article. Review.
-
Disruption of STAT3 signalling promotes KRAS-induced lung tumorigenesis.Nat Commun. 2015 Mar 3;6:6285. doi: 10.1038/ncomms7285. Nat Commun. 2015. PMID: 25734337 Free PMC article.
-
VEGFR2 survival and mitotic signaling depends on joint activation of associated C3ar1/C5ar1 and IL-6R-gp130.J Cell Sci. 2019 Mar 28;132(6):jcs219352. doi: 10.1242/jcs.219352. J Cell Sci. 2019. PMID: 30765465 Free PMC article.
-
17β-estradiol upregulates IL6 expression through the ERβ pathway to promote lung adenocarcinoma progression.J Exp Clin Cancer Res. 2018 Jul 3;37(1):133. doi: 10.1186/s13046-018-0804-5. J Exp Clin Cancer Res. 2018. PMID: 29970138 Free PMC article.
-
EGFR-rich extracellular vesicles derived from highly metastatic nasopharyngeal carcinoma cells accelerate tumour metastasis through PI3K/AKT pathway-suppressed ROS.J Extracell Vesicles. 2020 Oct;10(1):e12003. doi: 10.1002/jev2.12003. Epub 2020 Oct 30. J Extracell Vesicles. 2020. PMID: 33304472 Free PMC article.
References
-
- Siegfried JM. 2001. Women and lung cancer: does oestrogen play a role? Lancet Oncol 2:506–513 - PubMed
-
- Lam WK. 2005. Lung cancer in Asian women—the environment and genes. Respirology 10:408–417 - PubMed
-
- Ger LP , Liou SH , Shen CY , Kao SJ , Chen KT. 1992. [Risk factors of lung cancer]. J Formos Med Assoc 91(Suppl 3):S222–S231 - PubMed
-
- Xu X , Li B , Wang L. 1994. Gender difference in smoking effects on adult pulmonary function. Eur Respir J 7:477–483 - PubMed
-
- Zang EA , Wynder EL. 1996. Differences in lung cancer risk between men and women: examination of the evidence. J Natl Cancer Inst 88:183–192 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

