Array-based genomic screening at diagnosis and during follow-up in chronic lymphocytic leukemia
- PMID: 21546498
- PMCID: PMC3148910
- DOI: 10.3324/haematol.2010.039768
Array-based genomic screening at diagnosis and during follow-up in chronic lymphocytic leukemia
Abstract
Background: High-resolution genomic microarrays enable simultaneous detection of copy-number aberrations such as the known recurrent aberrations in chronic lymphocytic leukemia [del(11q), del(13q), del(17p) and trisomy 12], and copy-number neutral loss of heterozygosity. Moreover, comparison of genomic profiles from sequential patients' samples allows detection of clonal evolution.
Design and methods: We screened samples from 369 patients with newly diagnosed chronic lymphocytic leukemia from a population-based cohort using 250K single nucleotide polymorphism-arrays. Clonal evolution was evaluated in 59 follow-up samples obtained after 5-9 years.
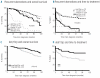
Results: At diagnosis, copy-number aberrations were identified in 90% of patients; 70% carried known recurrent alterations, including del(13q) (55%), trisomy 12 (10.5%), del(11q) (10%), and del(17p) (4%). Additional recurrent aberrations were detected on chromosomes 2 (1.9%), 4 (1.4%), 8 (1.6%) and 14 (1.6%). Thirteen patients (3.5%) displayed recurrent copy-number neutral loss of heterozygosity on 13q, of whom 11 had concurrent homozygous del(13q). Genomic complexity and large 13q deletions correlated with inferior outcome, while the former was linked to poor-prognostic aberrations. In the follow-up study, clonal evolution developed in 8/24 (33%) patients with unmutated IGHV, and in 4/25 (16%) IGHV-mutated and treated patients. In contrast, untreated patients with mutated IGHV (n=10) did not acquire additional aberrations. The most common secondary event, del(13q), was detected in 6/12 (50%) of all patients with acquired alterations. Interestingly, aberrations on, for example, chromosome 6q, 8p, 9p and 10q developed exclusively in patients with unmutated IGHV.
Conclusions: Whole-genome screening revealed a high frequency of genomic aberrations in newly diagnosed chronic lymphocytic leukemia. Clonal evolution was associated with other markers of aggressive disease and commonly included the known recurrent aberrations.
Figures



Similar articles
-
High-density screening reveals a different spectrum of genomic aberrations in chronic lymphocytic leukemia patients with 'stereotyped' IGHV3-21 and IGHV4-34 B-cell receptors.Haematologica. 2010 Sep;95(9):1519-25. doi: 10.3324/haematol.2009.021014. Epub 2010 Apr 26. Haematologica. 2010. PMID: 20421269 Free PMC article.
-
Importance of immunoglobulin heavy chain variable region mutational status in del(13q) chronic lymphocytic leukemia.Leuk Lymphoma. 2011 Oct;52(10):1873-81. doi: 10.3109/10428194.2011.585529. Epub 2011 Aug 18. Leuk Lymphoma. 2011. PMID: 21851216 Free PMC article.
-
Clinical, immunophenotypic, and molecular profiling of trisomy 12 in chronic lymphocytic leukemia and comparison with other karyotypic subgroups defined by cytogenetic analysis.Cancer Genet Cytogenet. 2006 Jul 15;168(2):109-19. doi: 10.1016/j.cancergencyto.2006.02.001. Cancer Genet Cytogenet. 2006. PMID: 16843100
-
The Heterogeneity of 13q Deletions in Chronic Lymphocytic Leukemia: Diagnostic Challenges and Clinical Implications.Genes (Basel). 2025 Feb 22;16(3):252. doi: 10.3390/genes16030252. Genes (Basel). 2025. PMID: 40149404 Free PMC article. Review.
-
Exploring the genetic landscape in chronic lymphocytic leukemia using high-resolution technologies.Leuk Lymphoma. 2013 Aug;54(8):1583-90. doi: 10.3109/10428194.2012.751530. Epub 2013 Feb 28. Leuk Lymphoma. 2013. PMID: 23167608 Review.
Cited by
-
Activation of the NOTCH1 pathway in chronic lymphocytic leukemia.Haematologica. 2012 Mar;97(3):328-30. doi: 10.3324/haematol.2012.061721. Haematologica. 2012. PMID: 22383743 Free PMC article. No abstract available.
-
Clonal evolution in chronic lymphocytic leukemia is associated with an unmutated IGHV status and frequently leads to a combination of loss of TP53 and TP53 mutation.Mol Biol Rep. 2022 Dec;49(12):12247-12252. doi: 10.1007/s11033-022-07888-y. Epub 2022 Sep 28. Mol Biol Rep. 2022. PMID: 36169893
-
Integrated mutational and cytogenetic analysis identifies new prognostic subgroups in chronic lymphocytic leukemia.Blood. 2013 Feb 21;121(8):1403-12. doi: 10.1182/blood-2012-09-458265. Epub 2012 Dec 13. Blood. 2013. PMID: 23243274 Free PMC article.
-
Characteristics of chronic lymphocytic leukemia in Senegal.BMC Hematol. 2016 Apr 23;16:10. doi: 10.1186/s12878-016-0051-y. eCollection 2016. BMC Hematol. 2016. PMID: 27110362 Free PMC article.
-
Monoallelic and biallelic deletions of 13q14 in a group of CLL/SLL patients investigated by CGH Haematological Cancer and SNP array (8x60K).Mol Cytogenet. 2016 Jan 6;9:1. doi: 10.1186/s13039-015-0212-x. eCollection 2016. Mol Cytogenet. 2016. PMID: 26740820 Free PMC article.
References
-
- Damle RN, Wasil T, Fais F, Ghiotto F, Valetto A, Allen SL, et al. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood. 1999;94(6):1840–7. - PubMed
-
- Dohner H, Stilgenbauer S, Benner A, Leupolt E, Krober A, Bullinger L, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343(26):1910–6. - PubMed
-
- Hamblin TJ, Davis Z, Gardiner A, Oscier DG, Stevenson FK. Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood. 1999;94(6):1848–54. - PubMed
-
- Juliusson G, Oscier DG, Fitchett M, Ross FM, Stockdill G, Mackie MJ, et al. Prognostic subgroups in B-cell chronic lymphocytic leukemia defined by specific chromosomal abnormalities. N Engl J Med. 1990;323(11):720–4. - PubMed
-
- Juliusson G, Robert KH, Ost A, Friberg K, Biberfeld P, Nilsson B, et al. Prognostic information from cytogenetic analysis in chronic B-lymphocytic leukemia and leukemic immunocytoma. Blood. 1985;65(1):134–41. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

