A region of the nucleosome required for multiple types of transcriptional silencing in Saccharomyces cerevisiae
- PMID: 21546544
- PMCID: PMC3176532
- DOI: 10.1534/genetics.111.129197
A region of the nucleosome required for multiple types of transcriptional silencing in Saccharomyces cerevisiae
Abstract
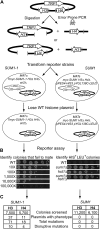
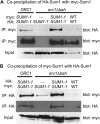
Extended heterochromatin domains, which are repressive to transcription and help define centromeres and telomeres, are formed through specific interactions between silencing proteins and nucleosomes. This study reveals that in Saccharomyces cerevisiae, the same nucleosomal surface is critical for the formation of multiple types of heterochromatin, but not for local repression mediated by a related transcriptional repressor. Thus, this region of the nucleosome may be generally important to long-range silencing. In S. cerevisiae, the Sir proteins perform long-range silencing, whereas the Sum1 complex acts locally to repress specific genes. A mutant form of Sum1p, Sum1-1p, achieves silencing in the absence of Sir proteins. A genetic screen identified mutations in histones H3 and H4 that disrupt Sum1-1 silencing and fall in regions of the nucleosome previously known to disrupt Sir silencing and rDNA silencing. In contrast, no mutations were identified that disrupt wild-type Sum1 repression. Mutations that disrupt silencing fall in two regions of the nucleosome, the tip of the H3 tail and a surface of the nucleosomal core (LRS domain) and the adjacent base of the H4 tail. The LRS/H4 tail region interacts with the Sir3p bromo-adjacent homology (BAH) domain to facilitate Sir silencing. By analogy, this study is consistent with the LRS/H4 tail region interacting with Orc1p, a paralog of Sir3p, to facilitate Sum1-1 silencing. Thus, the LRS/H4 tail region of the nucleosome may be relatively accessible and facilitate interactions between silencing proteins and nucleosomes to stabilize long-range silencing.
Figures




References
-
- Auth T., Kunkel E., Grummt F., 2006. Interaction between HP1alpha and replication proteins in mammalian cells. Exp. Cell Res. 312: 3349–3359 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

