SHAPE analysis of the FIV Leader RNA reveals a structural switch potentially controlling viral packaging and genome dimerization
- PMID: 21546549
- PMCID: PMC3159445
- DOI: 10.1093/nar/gkr252
SHAPE analysis of the FIV Leader RNA reveals a structural switch potentially controlling viral packaging and genome dimerization
Abstract
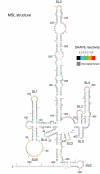
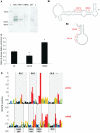
Feline immunodeficiency virus (FIV) infects many species of cat, and is related to HIV, causing a similar pathology. High-throughput selective 2' hydroxyl acylation analysed by primer extension (SHAPE), a technique that allows structural interrogation at each nucleotide, was used to map the secondary structure of the FIV packaging signal RNA. Previous studies of this RNA showed four conserved stem-loops, extensive long-range interactions (LRIs) and a small, palindromic stem-loop (SL5) within the gag open reading frame (ORF) that may act as a dimerization initiation site (DIS), enabling the virus to package two copies of its genome. Our analyses of wild-type (wt) and mutant RNAs suggest that although the four conserved stem-loops are static structures, the 5' and 3' regions previously shown to form LRI also adopt an alternative, yet similarly conserved conformation, in which the putative DIS is occluded, and which may thus favour translational and splicing functions over encapsidation. SHAPE and in vitro dimerization assays were used to examine SL5 mutants. Dimerization contacts appear to be made between palindromic loop sequences in SL5. As this stem-loop is located within the gag ORF, recognition of a dimeric RNA provides a possible mechanism for the specific packaging of genomic over spliced viral RNAs.
Figures





References
-
- Yamamoto JK, Sparger E, Ho EW, Andersen PR, O'Connor TP, Mandell CP, Lowenstine L, Munn R, Pedersen NC. Pathogenesis of experimentally induced feline immunodeficiency virus infection in cats. Am. J. Vet. Res. 1988;49:1246–1258. - PubMed
-
- Pedersen NC, Ho EW, Brown ML, Yamamoto JK. Isolation of a T-lymphotropic virus from domestic cats with an immunodeficiency-like syndrome. Science. 1987;235:790–793. - PubMed
-
- Siebelink KH, Chu IH, Rimmelzwaan GF, Weijer K, van Herwijnen R, Knell P, Egberink HF, Bosch ML, Osterhaus AD. Feline immunodeficiency virus (FIV) infection in the cat as a model for HIV infection in man: FIV-induced impairment of immune function. AIDS Res. Hum. Retroviruses. 1990;6:1373–1378. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

