Widespread expression of piRNA-like molecules in somatic tissues
- PMID: 21546553
- PMCID: PMC3159465
- DOI: 10.1093/nar/gkr298
Widespread expression of piRNA-like molecules in somatic tissues
Abstract
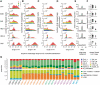
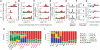
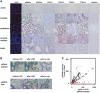
Piwi-interacting RNA (piRNA) are small RNA abundant in the germline across animal species. In fruit flies and mice, piRNA have been implicated in maintenance of genomic integrity by transposable elements silencing. Outside of the germline, piRNA have only been found in fruit fly ovarian follicle cells. Previous studies have further reported presence of multiple piRNA-like small RNA (pilRNA) in fly heads and a small number of pilRNA have been reported in mouse tissues and in human NK cells. Here, we analyze high-throughput small RNA sequencing data in more than 130 fruit fly, mouse and rhesus macaque samples. The results show widespread presence of pilRNA, displaying all known characteristics of piRNA in multiple somatic tissues of these three species. In mouse pancreas and macaque epididymis, pilRNA abundance was compatible with piRNA abundance in the germline. Using in situ hybridizations, we further demonstrate pilRNA co-localization with mRNA expression of Piwi-family genes in all macaque tissues. Further, using western blot, we have shown the expression of Miwi protein in mouse pancreas. These findings indicate that piRNA-like molecules might play important roles outside of the germline.
Figures

References
-
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. - PubMed
-
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. - PubMed
-
- Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

