The chloroplast genome of the green alga Schizomeris leibleinii (Chlorophyceae) provides evidence for bidirectional DNA replication from a single origin in the chaetophorales
- PMID: 21546564
- PMCID: PMC3138424
- DOI: 10.1093/gbe/evr037
The chloroplast genome of the green alga Schizomeris leibleinii (Chlorophyceae) provides evidence for bidirectional DNA replication from a single origin in the chaetophorales
Abstract
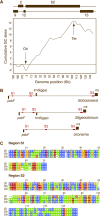
In the Chlorophyceae, the chloroplast genome is extraordinarily fluid in architecture and displays unique features relative to other groups of green algae. For the Chaetophorales, 1 of the 5 major lineages of the Chlorophyceae, it has been shown that the distinctive architecture of the 223,902-bp genome of Stigeoclonium helveticum is consistent with bidirectional DNA replication from a single origin. Here, we report the 182,759-bp chloroplast genome sequence of Schizomeris leibleinii, a member of the earliest diverging lineage of the Chaetophorales. Like its Stigeoclonium homolog, the Schizomeris genome lacks a large inverted repeat encoding the rRNA operon and displays a striking bias in coding regions that is associated with a bias in base composition along each strand. Our results support the notion that these two chaetophoralean genomes replicate bidirectionally from a putative origin located in the vicinity of the small subunit ribosomal RNA gene. Their shared structural characteristics were most probably inherited from the common ancestor of all chaetophoralean algae. Short dispersed repeats account for most of the 41-kb size variation between the Schizomeris and Stigeoclonium genomes, and there is no indication that homologous recombination between these repeated elements led to the observed gene rearrangements. A comparison of the extent of variation sustained by the Stigeoclonium and Schizomeris chloroplast DNAs (cpDNAs) with that observed for the cpDNAs of the chlamydomonadalean Chlamydomonas and Volvox suggests that gene rearrangements as well as changes in the abundance of intergenic and intron sequences occurred at a slower pace in the Chaetophorales than in the Chlamydomonadales.
Figures

Similar articles
-
Chloroplast DNA sequence of the green alga Oedogonium cardiacum (Chlorophyceae): unique genome architecture, derived characters shared with the Chaetophorales and novel genes acquired through horizontal transfer.BMC Genomics. 2008 Jun 16;9:290. doi: 10.1186/1471-2164-9-290. BMC Genomics. 2008. PMID: 18558012 Free PMC article.
-
The exceptionally large chloroplast genome of the green alga Floydiella terrestris illuminates the evolutionary history of the Chlorophyceae.Genome Biol Evol. 2010 Jul 12;2:240-56. doi: 10.1093/gbe/evq014. Genome Biol Evol. 2010. PMID: 20624729 Free PMC article.
-
Distinctive architecture of the chloroplast genome in the chlorophycean green alga Stigeoclonium helveticum.Mol Genet Genomics. 2006 Nov;276(5):464-77. doi: 10.1007/s00438-006-0156-2. Epub 2006 Aug 31. Mol Genet Genomics. 2006. PMID: 16944205
-
The complete chloroplast DNA sequence of the green alga Oltmannsiellopsis viridis reveals a distinctive quadripartite architecture in the chloroplast genome of early diverging ulvophytes.BMC Biol. 2006 Feb 3;4:3. doi: 10.1186/1741-7007-4-3. BMC Biol. 2006. PMID: 16472375 Free PMC article.
-
Hydrocytium expands the phylogenetic, morphological, and genomic diversity of the poorly known green algal order Chaetopeltidales.Am J Bot. 2023 Nov;110(11):e16238. doi: 10.1002/ajb2.16238. Epub 2023 Oct 24. Am J Bot. 2023. PMID: 37661934 Review.
Cited by
-
The chloroplast genomes of Bryopsis plumosa and Tydemania expeditiones (Bryopsidales, Chlorophyta): compact genomes and genes of bacterial origin.BMC Genomics. 2015 Mar 17;16(1):204. doi: 10.1186/s12864-015-1418-3. BMC Genomics. 2015. PMID: 25879186 Free PMC article.
-
Dynamic Evolution of the Chloroplast Genome in the Green Algal Classes Pedinophyceae and Trebouxiophyceae.Genome Biol Evol. 2015 Jul 1;7(7):2062-82. doi: 10.1093/gbe/evv130. Genome Biol Evol. 2015. PMID: 26139832 Free PMC article.
-
Distinctive Architecture of the Chloroplast Genome in the Chlorodendrophycean Green Algae Scherffelia dubia and Tetraselmis sp. CCMP 881.PLoS One. 2016 Feb 5;11(2):e0148934. doi: 10.1371/journal.pone.0148934. eCollection 2016. PLoS One. 2016. PMID: 26849226 Free PMC article.
-
Taxonomic scheme of the order Chaetophorales (Chlorophyceae, Chlorophyta) based on chloroplast genomes.BMC Genomics. 2020 Jun 26;21(1):442. doi: 10.1186/s12864-020-06845-y. BMC Genomics. 2020. PMID: 32590931 Free PMC article.
-
Proliferation of group II introns in the chloroplast genome of the green alga Oedocladium carolinianum (Chlorophyceae).PeerJ. 2016 Oct 25;4:e2627. doi: 10.7717/peerj.2627. eCollection 2016. PeerJ. 2016. PMID: 27812423 Free PMC article.
References
-
- Andersen RA, Berges JA, Harrison PJ, Watanabe MM. Appendix A—recipes for freshwater and seawater media. In: Andersen RA, editor. Algal culturing techniques. Burlington (MA): Elsevier Academic Press; 2005. pp. 429–538.
-
- Bélanger A-S, et al. Distinctive architecture of the chloroplast genome in the chlorophycean green alga Stigeoclonium helveticum. Mol Gen Genomics. 2006;276:464–477. - PubMed
-
- Boudreau E, Turmel M. Gene rearrangements in Chlamydomonas chloroplast DNAs are accounted for by inversions and by the expansion/contraction of the inverted repeat. Plant Mol Biol. 1995;27:351–364. - PubMed
-
- Boudreau E, Turmel M. Extensive gene rearrangements in the chloroplast DNAs of Chlamydomonas species featuring multiple dispersed repeats. Mol Biol Evol. 1996;13:233–243. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

