Pro-apoptotic activity of inhibitory PAS domain protein (IPAS), a negative regulator of HIF-1, through binding to pro-survival Bcl-2 family proteins
- PMID: 21546903
- PMCID: PMC3190112
- DOI: 10.1038/cdd.2011.47
Pro-apoptotic activity of inhibitory PAS domain protein (IPAS), a negative regulator of HIF-1, through binding to pro-survival Bcl-2 family proteins
Abstract
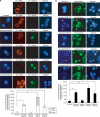
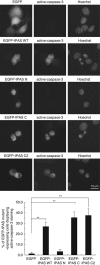
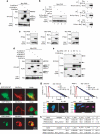
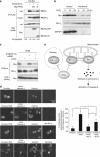
Inhibitory PAS (Per/Arnt/Sim) domain protein (IPAS) is a dominant negative transcription factor that represses hypoxia-inducible factor 1 (HIF-1) activity. In this study, we show that IPAS also functions as a pro-apoptotic protein through binding to pro-survival Bcl-2 family members. In a previous paper, we reported that NF-κB-dependent IPAS induction by cobalt chloride repressed the hypoxic response in PC12 cells. We found that prolonged incubation under the same conditions caused apoptosis in PC12 cells. Repression of IPAS induction protected cells from apoptosis. Furthermore, knockdown of IPAS recovered cell viability. EGFP-IPAS protein was localized in both the nucleus and the cytoplasm, with a large fraction associated with mitochondria. Mitochondrial IPAS induced mitochondria depolarization and caspase-3 activation. Immunoprecipitation assays revealed that IPAS is associated with Bcl-x(L), Bcl-w and Mcl-1. The association of IPAS with Bcl-x(L) was also observed in living cells by the FLIM-based FRET analysis, indicating direct binding between the two proteins. IPAS contributed to dysfunction of Bcl-x(L) by inhibiting the interaction of Bcl-x(L) with Bax. These results demonstrate that IPAS functions as a dual function protein involved in transcription repression and apoptosis.
Figures






References
-
- Hirota K, Semenza GL. Regulation of hypoxia-inducible factor 1 by prolyl and asparaginyl hydroxylases. Biochem Biophys Res Commun. 2005;338:610–616. - PubMed
-
- Kaelin WG., Jr The von Hippel-Lindau protein, HIF hydroxylation, and oxygen sensing. Biochem Biophys Res Commun. 2005;338:627–638. - PubMed
-
- Schofield CJ, Ratcliffe PJ. Signalling hypoxia by HIF hydroxylases. Biochem Biophys Res Commun. 2005;338:617–626. - PubMed
-
- Makino Y, Cao R, Svensson K, Bertilsson G, Asman M, Tanaka H, et al. Inhibitory PAS domain protein is a negative regulator of hypoxia-inducible gene expression. Nature. 2001;414:550–554. - PubMed
-
- Makino Y, Uenishi R, Okamoto K, Isoe T, Hosono O, Tanaka H, et al. Transcriptional up-regulation of inhibitory PAS domain protein gene expression by hypoxia-inducible factor 1 (HIF-1): a negative feedback regulatory circuit in HIF-1-mediated signaling in hypoxic cells. J Biol Chem. 2007;282:14073–14082. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

