Alphavirus Entry and Membrane Fusion
- PMID: 21546978
- PMCID: PMC3086016
- DOI: 10.3390/v2040796
Alphavirus Entry and Membrane Fusion
Abstract
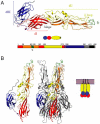
The study of enveloped animal viruses has greatly advanced our understanding of the general properties of membrane fusion and of the specific pathways that viruses use to infect the host cell. The membrane fusion proteins of the alphaviruses and flaviviruses have many similarities in structure and function. As reviewed here, alphaviruses use receptor-mediated endocytic uptake and low pH-triggered membrane fusion to deliver their RNA genomes into the cytoplasm. Recent advances in understanding the biochemistry and structure of the alphavirus membrane fusion protein provide a clearer picture of this fusion reaction, including the protein's conformational changes during fusion and the identification of key domains. These insights into the alphavirus fusion mechanism suggest new areas for experimental investigation and potential inhibitor strategies for anti-viral therapy.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
