High-flux isobutanol production using engineered Escherichia coli: a bioreactor study with in situ product removal
- PMID: 21547458
- PMCID: PMC3094657
- DOI: 10.1007/s00253-011-3173-y
High-flux isobutanol production using engineered Escherichia coli: a bioreactor study with in situ product removal
Abstract
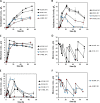
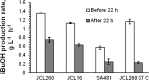
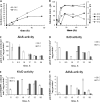
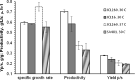
Promising approaches to produce higher alcohols, e.g., isobutanol, using Escherichia coli have been developed with successful results. Here, we translated the isobutanol process from shake flasks to a 1-L bioreactor in order to characterize three E. coli strains. With in situ isobutanol removal from the bioreactor using gas stripping, the engineered E. coli strain (JCL260) produced more than 50 g/L in 72 h. In addition, the isobutanol production by the parental strain (JCL16) and the high isobutanol-tolerant mutant (SA481) were compared with JCL260. Interestingly, we found that the isobutanol-tolerant strain in fact produced worse than either JCL16 or JCL260. This result suggests that in situ product removal can properly overcome isobutanol toxicity in E. coli cultures. The isobutanol productivity was approximately twofold and the titer was 9% higher than n-butanol produced by Clostridium in a similar integrated system.
Figures



References
-
- Alper H, Moxley J, Nevoigt E, Fink GR, Stephanopoulos G. Engineering yeast transcription machinery for improved ethanol tolerance and production. Science. 2006;314:1565–1568. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

