STIM proteins and the endoplasmic reticulum-plasma membrane junctions
- PMID: 21548779
- PMCID: PMC3897197
- DOI: 10.1146/annurev-biochem-061609-165311
STIM proteins and the endoplasmic reticulum-plasma membrane junctions
Abstract
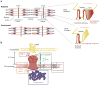
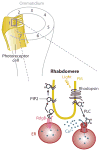
Eukaryotic organelles can interact with each other through stable junctions where the two membranes are kept in close apposition. The junction that connects the endoplasmic reticulum to the plasma membrane (ER-PM junction) is unique in providing a direct communication link between the ER and the PM. In a recently discovered signaling process, STIM (stromal-interacting molecule) proteins sense a drop in ER Ca(2+) levels and directly activate Orai PM Ca(2+) channels across the junction space. In an inverse process, a voltage-gated PM Ca(2+) channel can directly open ER ryanodine-receptor Ca(2+) channels in striated-muscle cells. Although ER-PM junctions were first described 50 years ago, their broad importance in Ca(2+) signaling, as well as in the regulation of cholesterol and phosphatidylinositol lipid transfer, has only recently been realized. Here, we discuss research from different fields to provide a broad perspective on the structures and unique roles of ER-PM junctions in controlling signaling and metabolic processes.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

