The four Zn fingers of MBNL1 provide a flexible platform for recognition of its RNA binding elements
- PMID: 21548961
- PMCID: PMC3103431
- DOI: 10.1186/1471-2199-12-20
The four Zn fingers of MBNL1 provide a flexible platform for recognition of its RNA binding elements
Abstract
Background: Muscleblind-like 1 (MBNL1) is an alternative splicing factor containing four CCCH Zinc fingers (ZnFs). The sequestration of MBNL1 by expanded CUG and CCUG repeats is a major component in causing myotonic dystrophy. In addition to binding the structured expanded CUG and CCUG repeats; previous results suggested that MBNL1 binds single-stranded RNAs containing GC dinucleotides.
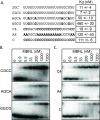
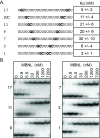
Results: We performed a systematic analysis of MBNL1 binding to single-stranded RNAs. These studies revealed that a single GC dinucleotide in poly-uridine is sufficient for MBNL1 binding and that a second GC dinucleotide confers higher affinity MBNL1 binding. However additional GC dinucleotides do not enhance RNA binding. We also showed that the RNA sequences adjacent to the GC dinucleotides play an important role in MBNL1 binding with the following preference: uridines >cytidines >adenosines >guanosines. For high affinity binding by MBNL1, the distance between the two GC dinucleotides can vary from 1 to 17 nucleotides.
Conclusions: These results suggest that MBNL1 is highly flexible and able to adopt different conformations to recognize RNAs with varying sequence configurations. Although MBNL1 contains four ZnFs, only two ZnF - GC dinucleotide interactions are necessary for high affinity binding.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

