The molecular and cellular basis of variable craniofacial phenotypes and their genetic rescue in Twisted gastrulation mutant mice
- PMID: 21549111
- PMCID: PMC3105466
- DOI: 10.1016/j.ydbio.2011.04.026
The molecular and cellular basis of variable craniofacial phenotypes and their genetic rescue in Twisted gastrulation mutant mice
Abstract
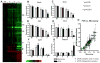
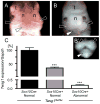
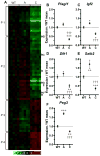
The severity of numerous developmental abnormalities can vary widely despite shared genetic causes. Mice deficient in Twisted gastrulation (Twsg1(-/-)) display such phenotypic variation, developing a wide range of craniofacial malformations on an isogenic C57BL/6 strain background. To examine the molecular basis for this reduced penetrance and variable expressivity, we used exon microarrays to analyze gene expression in mandibular arches from several distinct, morphologically defined classes of Twsg1(-/-) and wild type (WT) embryos. Hierarchical clustering analysis of transcript levels identified numerous differentially expressed genes, clearly distinguishing severely affected and unaffected Twsg1(-/-) mutants from WT embryos. Several genes that play well-known roles in craniofacial development were upregulated in unaffected Twsg1(-/-) mutant embryos, suggesting that they may compensate for the loss of TWSG1. Imprinted genes were overrepresented among genes that were differentially expressed particularly between affected and unaffected mutants. The most severely affected embryos demonstrated increased p53 signaling and increased expression of its target, Trp53inp1. The frequency of craniofacial defects significantly decreased with a reduction of p53 gene dosage from 44% in Twsg1(-/-)p53(+/+) pups (N=675) to 30% in Twsg1(-/-)p53(+/-) (N=47, p=0.04) and 15% in Twsg1(-/-)p53(-/-) littermates (N=39, p=0.001). In summary, these results demonstrate that phenotypic variability in Twsg1(-/-) mice is associated with differential expression of certain developmentally regulated genes, and that craniofacial defects can be partially rescued by reduced p53 levels. We postulate that variable responses to stress may contribute to variable craniofacial phenotypes by triggering differential expression of genes and variable cellular apoptosis.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures




References
-
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. Series B 1995
-
- Berdal A, Molla M, Hotton D, Aïoub M, Lézot F, Néfussi JR, Goubin G. Differential impact of MSX1 and MSX2 homeogenes on mouse maxillofacial skeleton. Cells, tissues, organs. 2009;189:126–132. - PubMed
-
- Bobola N, Carapuco M, Ohnemus S, Kanzler B, Leibbrandt A, Neubuser A, Drouin J, Mallo M. Mesenchymal patterning by Hoxa2 requires blocking Fgf-dependent activation of Ptx1. Development. 2003;130:3403–3414. - PubMed
-
- Braybrook C, Doudney K, Marcano AC, Arnason A, Bjornsson A, Patton MA, Goodfellow PJ, Moore GE, Stanier P. The T-box transcription factor gene TBX22 is mutated in X-linked cleft palate and ankyloglossia. Nat Genet. 2001;29:179–183. - PubMed
-
- Bruce S, Hannula-Jouppi K, Peltonen J, Kere J, Lipsanen-Nyman M. Clinically Distinct Epigenetic Subgroups in Silver-Russell Syndrome: The Degree of H19 Hypomethylation Associates with Phenotype Severity and Genital and Skeletal Anomalies. Journal of Clinical Endocrinology & Metabolism. 2009;94:579–587. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

