Interference of ribosomal frameshifting by antisense peptide nucleic acids suppresses SARS coronavirus replication
- PMID: 21549154
- PMCID: PMC4728714
- DOI: 10.1016/j.antiviral.2011.04.009
Interference of ribosomal frameshifting by antisense peptide nucleic acids suppresses SARS coronavirus replication
Abstract
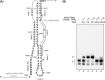
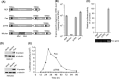
The programmed -1 ribosomal frameshifting (-1 PRF) utilized by eukaryotic RNA viruses plays a crucial role for the controlled, limited synthesis of viral RNA replicase polyproteins required for genome replication. The viral RNA replicase polyproteins of severe acute respiratory syndrome coronavirus (SARS-CoV) are encoded by the two overlapping open reading frames 1a and 1b, which are connected by a -1 PRF signal. We evaluated the antiviral effects of antisense peptide nucleic acids (PNAs) targeting a highly conserved RNA sequence on the - PRF signal. The ribosomal frameshifting was inhibited by the PNA, which bound sequence-specifically a pseudoknot structure in the -1 PRF signal, in cell lines as assessed using a dual luciferase-based reporter plasmid containing the -1 PRF signal. Treatment of cells, which were transfected with a SARS-CoV-replicon expressing firefly luciferase, with the PNA fused to a cell-penetrating peptide (CPP) resulted in suppression of the replication of the SARS-CoV replicon, with a 50% inhibitory concentration of 4.4μM. There was no induction of type I interferon responses by PNA treatment, suggesting that the effect of PNA is not due to innate immune responses. Our results demonstrate that -1 PRF, critical for SARS-CoV viral replication, can be inhibited by CPP-PNA, providing an effective antisense strategy for blocking -1 PRF signals.
Copyright © 2011 Elsevier B.V. All rights reserved.
Figures

References
-
- Almazan F., Dediego M.L., Galan C., Escors D., Alvarez E., Ortego J., Sola I., Zuniga S., Alonso S., Moreno J.L., Nogales A., Capiscol C., Enjuanes L. Construction of a severe acute respiratory syndrome coronavirus infectious cDNA clone and a replicon to study coronavirus RNA synthesis. J. Virol. 2006;80:10900–10906. - PMC - PubMed
-
- Baranov P.V., Gesteland R.F., Atkins J.F. Recoding: translational bifurcations in gene expression. Gene. 2002;286:187–201. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

