Single-molecule fluorescence measurements of ribosomal translocation dynamics
- PMID: 21549313
- PMCID: PMC3090999
- DOI: 10.1016/j.molcel.2011.03.024
Single-molecule fluorescence measurements of ribosomal translocation dynamics
Abstract
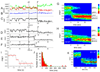
We employ single-molecule fluorescence resonance energy transfer (smFRET) to study structural dynamics over the first two elongation cycles of protein synthesis, using ribosomes containing either Cy3-labeled ribosomal protein L11 and A- or P-site Cy5-labeled tRNA or Cy3- and Cy5-labeled tRNAs. Pretranslocation (PRE) complexes demonstrate fluctuations between classical and hybrid forms, with concerted motions of tRNAs away from L11 and from each other when classical complex converts to hybrid complex. EF-G⋅GTP binding to both hybrid and classical PRE complexes halts these fluctuations prior to catalyzing translocation to form the posttranslocation (POST) complex. EF-G dependent translocation from the classical PRE complex proceeds via transient formation of a short-lived hybrid intermediate. A-site binding of either EF-G to the PRE complex or of aminoacyl-tRNA⋅EF-Tu ternary complex to the POST complex markedly suppresses ribosome conformational lability.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures
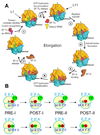
 colored letters M, R and F indicate mRNA codons for fMet, Arg and Phe. The black letters M, R, and F refer to tRNAfMet, tRNAArg and tRNAPhe.
colored letters M, R and F indicate mRNA codons for fMet, Arg and Phe. The black letters M, R, and F refer to tRNAfMet, tRNAArg and tRNAPhe.  dots indicate Cy5 labeling of the tRNAs.
dots indicate Cy5 labeling of the tRNAs.  dots indicate Cy3 labeling of L11 or tRNA in Lt or tt complexes, respectively.
dots indicate Cy3 labeling of L11 or tRNA in Lt or tt complexes, respectively.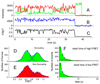


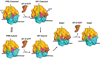
References
-
- Blanchard SC, Gonzalez RL, Kim HD, Chu S, Puglisi JD. tRNA selection and kinetic proofreading in translation. Nat Struct Mol Biol. 2004a;11:1008–1014. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

