Diverse interactions of retroviral Gag proteins with RNAs
- PMID: 21550256
- PMCID: PMC3130074
- DOI: 10.1016/j.tibs.2011.04.001
Diverse interactions of retroviral Gag proteins with RNAs
Abstract
Retrovirus particles are constructed from a single virus-encoded protein, termed Gag. Given that assembly is an essential step in the viral replication cycle, it is a potential target for antiviral therapy. However, such an approach has not yet been exploited because of the lack of fundamental knowledge concerning the structures and interactions responsible for assembly. Assembling an infectious particle entails a remarkably diverse array of interactions, both specific and nonspecific, between Gag proteins and RNAs. These interactions are essential for the construction of the particle, for packaging of the viral RNA into the particle, and for placement of the primer for viral DNA synthesis. Recent results have provided some new insights into each of these interactions. In the case of HIV-1 Gag, it is clear that more than one domain of the protein contributes to Gag-RNA interaction.
Published by Elsevier Ltd.
Figures

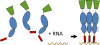

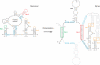

References
-
- Coffin JM, et al. Retroviruses. Cold Spring Harbor Laboratory Press; 1997. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

