Significant alteration of gene expression in wood decay fungi Postia placenta and Phanerochaete chrysosporium by plant species
- PMID: 21551287
- PMCID: PMC3127733
- DOI: 10.1128/AEM.00508-11
Significant alteration of gene expression in wood decay fungi Postia placenta and Phanerochaete chrysosporium by plant species
Abstract
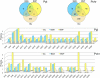
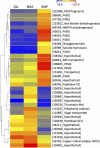
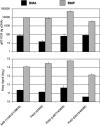
Identification of specific genes and enzymes involved in conversion of lignocellulosics from an expanding number of potential feedstocks is of growing interest to bioenergy process development. The basidiomycetous wood decay fungi Phanerochaete chrysosporium and Postia placenta are promising in this regard because they are able to utilize a wide range of simple and complex carbon compounds. However, systematic comparative studies with different woody substrates have not been reported. To address this issue, we examined gene expression of these fungi colonizing aspen (Populus grandidentata) and pine (Pinus strobus). Transcript levels of genes encoding extracellular glycoside hydrolases, thought to be important for hydrolytic cleavage of hemicelluloses and cellulose, showed little difference for P. placenta colonizing pine versus aspen as the sole carbon source. However, 164 genes exhibited significant differences in transcript accumulation for these substrates. Among these, 15 cytochrome P450s were upregulated in pine relative to aspen. Of 72 P. placenta extracellular proteins identified unambiguously by mass spectrometry, 52 were detected while colonizing both substrates and 10 were identified in pine but not aspen cultures. Most of the 178 P. chrysosporium glycoside hydrolase genes showed similar transcript levels on both substrates, but 13 accumulated >2-fold higher levels on aspen than on pine. Of 118 confidently identified proteins, 31 were identified in both substrates and 57 were identified in pine but not aspen cultures. Thus, P. placenta and P. chrysosporium gene expression patterns are influenced substantially by wood species. Such adaptations to the carbon source may also reflect fundamental differences in the mechanisms by which these fungi attack plant cell walls.
Figures
References
-
- Adler E. 1977. Lignin chemistry. Past, present and future. Wood Sci. Technol. 11:169–218
-
- Baldrian P., Valaskova V. 2008. Degradation of cellulose by basidiomycetous fungi. FEMS Microbiol. Rev. 32:501–521 - PubMed
-
- Banerjee G., Scott-Craig J. S., Walton J. D. 2010. Improving enzymes for biomass conversion: a basic research perspective. Bioenerg. Res. 3:82–92
-
- Bao W., Lymar E., Renganathan V. 1994. Optimization of cellobiose dehydrogenase and β-glucosidase production by cellulose-degrading cultures of Phanerochaete chrysosporium. Appl. Biochem. Biotechnol. 42:642–646
-
- Benjamini Y., Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57:289–300
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

