The lysis-lysogeny decision of bacteriophage 933W: a 933W repressor-mediated long-distance loop has no role in regulating 933W P(RM) activity
- PMID: 21551291
- PMCID: PMC3133280
- DOI: 10.1128/JB.00119-11
The lysis-lysogeny decision of bacteriophage 933W: a 933W repressor-mediated long-distance loop has no role in regulating 933W P(RM) activity
Abstract
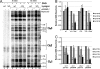
Our data show that unlike bacteriophage λ, repressor bound at O(L) of bacteriophage 933W has no role in regulation of 933W repressor occupancy of 933W O(R)3 or the transcriptional activity of 933W P(RM). This finding suggests that a cooperative long-range loop between repressor tetramers bound at O(R) and O(L) does not form in bacteriophage 933W. Nonetheless, 933W forms lysogens, and 933W prophage display a threshold response to UV induction similar to related lambdoid phages. Hence, the long-range loop thought to be important for constructing a threshold response in lambdoid bacteriophages is dispensable. The lack of a loop requires bacteriophage 933W to use a novel strategy in regulating its lysis-lysogeny decisions. As part of this strategy, the difference between the repressor concentrations needed to bind O(R)2 and activate 933W P(RM) transcription or bind O(R)3 and repress transcription from P(RM) is <2-fold. Consequently, P(RM) is never fully activated, reaching only ∼25% of the maximum possible level of repressor-dependent activation before repressor-mediated repression occurs. The 933W repressor also apparently does not bind cooperatively to the individual sites in O(R) and O(L). This scenario explains how, in the absence of DNA looping, bacteriophage 933W displays a threshold effect in response to DNA damage and suggests how 933W lysogens behave as "hair triggers" with spontaneous induction occurring to a greater extent in this phage than in other lambdoid phages.
Figures




References
-
- Arber W., et al. 1983. Lambda II, p. 433–466 Cold Spring Harbor Laboratory, Cold Spring Harbor, NY
-
- Baek K., Svenningsen S., Eisen H., Sneppen K., Brown S. 2003. Single-cell analysis of lambda immunity regulation. J. Mol. Biol. 334:363–372 - PubMed
-
- Beckett D., Burz D. S., Ackers G. K., Sauer R. T. 1993. Isolation of lambda repressor mutants with defects in cooperative operator binding. Biochemistry 32:9073–9079 - PubMed
-
- Beckett D., Koblan K. S., Ackers G. K. 1991. Quantitative study of protein association at picomolar concentrations: the lambda phage cI repressor. Anal. Biochem. 196:69–75 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

