Proteomic analysis yields an unexpected trans-acting point in control of the human sympathochromaffin phenotype
- PMID: 21551321
- PMCID: PMC3319683
- DOI: 10.1161/CIRCGENETICS.110.957886
Proteomic analysis yields an unexpected trans-acting point in control of the human sympathochromaffin phenotype
Abstract
Background: The secretory protein chromogranin A (CHGA) plays a necessary role in formation of catecholamine storage vesicles and gives rise to a catecholamine release-inhibitory fragment. Because genetic variation in the proximal human CHGA promoter predicts autonomic function and blood pressure, we explored how a common genetic variant alters transcription of the gene.
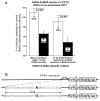
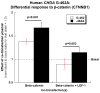
Methods and results: Bioinformatic analysis suggested that the common G-462A promoter variant (rs9658634) may disrupt as many as 3 transcriptional control motifs: LEF1, COUP-TF, and PPARγ-RXRα. During electrophoretic mobility shifts, chromaffin cell nuclear proteins bound specifically to the A (though not G) allele of CHGA promoter G-462A. On oligonucleotide affinity chromatography followed by electrospray ionization followed by 2-dimensional (tandem) mass spectrometry analysis of A allele eluates, the transcription factor LEF1 (lymphoid enhancer-binding factor-1) was identified. Interaction of LEF1 with the A allele at G-462A was confirmed by supershift. On cotransfection, LEF1 discriminated between the allelic variants, especially in chromaffin cells. Allele specificity of trans-activation by LEF1 was transferable to an isolated G-462A element fused to a heterologous (SV40) promoter. Because β-catenin (CTNNB1) can heterodimerize with LEF1, we tested the effect of cotransfection of this factor and again found A allele-specific perturbation of CHGA transcription.
Conclusions: Common genetic variation within the human CHGA promoter alters the interaction of specific factors in trans with the promoter, with LEF1 identified by proteomic analysis and confirmed by supershift. Coexpression experiments show functional effects of LEF1 and CTNNB1 on CHGA promoter. The findings document a novel role for components of the immune and WNT pathways in control of human sympathochromaffin phenotypes.
Conflict of interest statement
Figures




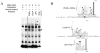
Similar articles
-
A haplotype variant of the human chromogranin A gene (CHGA) promoter increases CHGA expression and the risk for cardiometabolic disorders.J Biol Chem. 2017 Aug 25;292(34):13970-13985. doi: 10.1074/jbc.M117.778134. Epub 2017 Jun 30. J Biol Chem. 2017. PMID: 28667172 Free PMC article.
-
Genetic variation within a metabolic motif in the chromogranin a promoter: pleiotropic influence on cardiometabolic risk traits in twins.Am J Hypertens. 2012 Jan;25(1):29-40. doi: 10.1038/ajh.2011.163. Epub 2011 Sep 15. Am J Hypertens. 2012. PMID: 21918574 Free PMC article.
-
Common genetic variants in the chromogranin A promoter alter autonomic activity and blood pressure.Kidney Int. 2008 Jul;74(1):115-25. doi: 10.1038/ki.2008.113. Epub 2008 Apr 23. Kidney Int. 2008. PMID: 18432188 Free PMC article.
-
Heredity of endothelin secretion: human twin studies reveal the influence of polymorphism at the chromogranin A locus, a novel determinant of endothelial function.Circulation. 2007 May 1;115(17):2282-91. doi: 10.1161/CIRCULATIONAHA.106.648345. Epub 2007 Apr 16. Circulation. 2007. PMID: 17438153
-
Chromogranin A: a novel susceptibility gene for essential hypertension.Cell Mol Life Sci. 2010 Mar;67(6):861-74. doi: 10.1007/s00018-009-0208-y. Epub 2009 Nov 27. Cell Mol Life Sci. 2010. PMID: 19943077 Free PMC article. Review.
Cited by
-
A haplotype variant of the human chromogranin A gene (CHGA) promoter increases CHGA expression and the risk for cardiometabolic disorders.J Biol Chem. 2017 Aug 25;292(34):13970-13985. doi: 10.1074/jbc.M117.778134. Epub 2017 Jun 30. J Biol Chem. 2017. PMID: 28667172 Free PMC article.
-
Chromogranin A and its derived peptides: potential regulators of cholesterol homeostasis.Cell Mol Life Sci. 2023 Aug 29;80(9):271. doi: 10.1007/s00018-023-04908-3. Cell Mol Life Sci. 2023. PMID: 37642733 Free PMC article. Review.
References
-
- Taupenot L, Harper KL, O’Connor DT. The chromogranin-secretogranin family. N Engl J Med. 2003;348:1134–1149. - PubMed
-
- Kim T, Tao-Cheng JH, Eiden LE, Loh YP. Chromogranin A, an “on/off” switch controlling dense-core secretory granule biogenesis. Cell. 2001;106:499–509. - PubMed
-
- Takiyyuddin MA, Cervenka JH, Hsiao RJ, Barbosa JA, Parmer RJ, O’Connor DT. Chromogranin A. Storage and release in hypertension. Hypertension. 1990;15:237–246. - PubMed
-
- Aardal S, Helle KB, Elsayed S, Reed RK, Serck-Hanssen G. Vasostatins, comprising the N-terminal domain of chromogranin A, suppress tension in isolated human blood vessel segments. J Neuroendocrinol. 1993;5:405–412. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

